Abstract
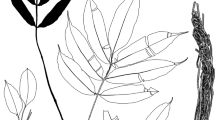
Disperis tomaszii (Orchidaceae, Orchidoideae) from Cameroon is described and illustrated in this study. The new species is morphologically similar to Disperis nitida and Disperis thomensis. Phylogenetic analyses of selected DNA regions (ITS, matK) indicate a possible relationship between the new species and Disperis anthoceros and Disperis dicerochila. The taxonomic affinity between D. tomaszii and the aforementioned species is discussed.







Similar content being viewed by others
References
BirdLife International (2010) BirdLife International—the information resource for BirdLife International. Available at: http://www.birdlife.org/. Accessed 25 Dec 2012
Brummitt RK, Powell CE (1992) Authors of plant names. A list of authors of scientific names of plants, with recommended standard form of their names including abbreviations. Royal Botanic Gardens, Kew
Chase MW, Hills HH (1991) Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon 40:215–220. https://doi.org/10.2307/1222975
Cuénoud P, Savolainen V, Chatrou LW, Powell M, Grayer RJ, Chase MW (2002) Molecular phylogenetics of Caryophyllales based on nuclear 18S rDNA and plastid rbcL, atpB and matK DNA sequences. Amer J Bot 89:132–144. https://doi.org/10.3732/ajb.89.1.132
Douzery E, Pridgeon AM, Kores PJ, Kurzwell H, Linder P, Chase MW (1999) Molecular phylogenetics of Diseae (Orchidaceae): a contribution from nuclear ribosomal ITS sequences. Amer J Bot 86:887–889
Edgar RC (2004) Muscle: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32:1792–1797
Erixon P, Svennblad B, Britton T, Oxelman B (2003) Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetic. Syst Biol 52:665–673. https://doi.org/10.1080/10635150390235485
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.2307/2408678
Fitch WM (1971) Towards defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416. https://doi.org/10.2307/2412116
Galtier N, Gouy M, Gautier C (1996) SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Bioinformatics 12:543–548. https://doi.org/10.1093/bioinformatics/12.6.543
Kores PJ, Molvray M, Weston PH, Hopper SD, Brown AP, Cameron KM, Chase MW (2001) A phylogenetic analysis of Diurideae (Orchidaceae) based on plastid DNA sequence data. Amer J Bot 88:1903–1914. https://doi.org/10.2307/3558366
Kurzweil H, Manning JC (2005) A synopsis of the genus Disperis Sw. (Orchidaceae). Adansonia 27:155–207
Manning JC, Linder HP (1992) Pollinators and evolution in Disperis (Orchidaceae), or why are there so many species? S Afr J Sci 88:38–49
Molvray M, Kores PJ, Chase MW (2000) Polyphyly of mycoheterotropic orchids and functional influences on floral and molecular characters. In: Morrison DA, Wilson KL (eds) Moncots: systematic and evolution. CSIRO Publishing, Collingwood, pp 441–448
Nylander JAA (2004) MrModeltest v2. Program distributed by the author, Evolutionary Biology Centre, Uppsala University, Uppsala
Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (2001) Genera Orchidacearum. Vol. 2: Orchidoideae (Part 1). Oxford University Press, New York
Ronquist H, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Schlechter R (1898) Monographie der Disperideae. Bull Herb Boissier 6:1–94
Swartz O (1800) Orchidernesslagterochacterupstallde. Kongl Vetensk Acad Nya Handl 21:202–254
Swofford DL (2000) PAUP*: Phylogenetic Analysis Using Parsymony (*and Other Methods). Version 4.0b2. Sinauer Associates, Sunderland
Szlachetko DL, Olszewski TS (1998) Orchidacees. In: Morat P (ed) Flore du Cameroun, Musèum National d’Histoire Naturalle, vol. 1. Yaounde, Paris, pp 1–327
Szlachetko DL, Rutkowski P (2000) Gynostemia Orchidalium 1. Apostasiaceae, Cypripediaceae, Orchidaceae (Thelymitroideae to Vanilloideae). Acta Bot Fenn 169:1–380
Szlachetko DL, Mytnik-Ejsmont J, Kras M, Rutkowski P, Baranow P, Górniak M (2010) Orchidaceae of West-Central Africa, vol. 1. Gdansk University Press, Gdansk
Thiers B (2016) Index Herbariorum: A global directory of public herbaria and associated staff. New York Botanical Garden, Bronx. http://sweetgum.nybg.org/science/ih/
Acknowledgements
We wish to thank Dr Przemysław Baranow and Dr Hanna B. Margońska for preparing drawings for this article.
Funding
This work was supported by a grant of the Ministry of Science and Higher Education [N N303 343735].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling editor: Ricarda Riina.
Electronic supplementary material
Below is the link to the electronic supplementary material.
606_2017_1471_MOESM3_ESM.pdf
Online Resource 3. Strict consensus tree obtained in maximum parsimony analysis of ITS1-5.8S-ITS2 sequences from species of Disperis (PDF 25 kb)
606_2017_1471_MOESM4_ESM.pdf
Online Resource 4. Strict consensus tree obtained in maximum parsimony analysis of part of matK gene sequences from species of Disperis (PDF 1357 kb)
606_2017_1471_MOESM5_ESM.pdf
Online Resource 5. Majority-rule consensus tree obtained in Bayesian analysis of part of matK gene sequences from species of Disperis. Values above branches represent posterior probabilities (≥ 50%) and bootstrap support (≥ 50%) from 1000 replicates (PP/BP) (PDF 1361 kb)
Information on Electronic supplementary material
Information on Electronic supplementary material
Online Resource 1. ITS data matrix used in the study.
Online Resource 2. matK data matrix used in the study.
Online Resource 3. Strict consensus tree obtained in maximum parsimony analysis of ITS1-5.8S-ITS2 sequences from species of Disperis.
Online Resource 4. Strict consensus tree obtained in maximum parsimony analysis of part of matK gene sequences from species of Disperis.
Online Resource 5. Majority-rule consensus tree obtained in Bayesian analysis of part of matK gene sequences from species of Disperis. Values above branches represent posterior probabilities (≥ 50%) and bootstrap support (≥ 50%) from 1000 replicates (PP/BP).
Rights and permissions
About this article
Cite this article
Szlachetko, D.L., Grochocka, E., Dudek, M. et al. Disperis tomaszii (Orchidaceae, Orchidoideae), a new species from Cameroon. Plant Syst Evol 304, 231–243 (2018). https://doi.org/10.1007/s00606-017-1471-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-017-1471-2