Abstract
The genus Ivodea is endemic to Madagascar and the Comoros and consists of 30 species. This study is the first to include the genus in a molecular phylogenetic analysis. We sequenced the plastid trnL–trnF and the nuclear ITS regions for three Ivodea species and revealed that the genus is monophyletic and most closely related to the African and Malagasy Vepris, refuting earlier suggestions of a close relationship between Ivodea and the Asian, Malesian, Australasian and Pacific genera Euodia and Melicope. Ivodea and Vepris provide another example of closely related pairs of Rutaceous groups that have drupaceous and capsular/follicular fruits, respectively, thus further confirming that fruit types are not suited to delimit subfamilies in Rutaceae, as has often been done in the past. Ivodea was the last of the seven Malagasy genera to be included in the Rutaceae phylogeny, making it possible to conduct an assessment of biogeographic affinities of the genera that occur on the island. Our assessments based on sister-group relationships suggest that the eight lineages (representing seven genera) of Malagasy Rutaceae either have African or have Asian affinities. Two lineages have an African origin, and one lineage has an Asian origin. Taxon sampling is insufficient to interpret the directionality of dispersal events in the remaining lineages.
Similar content being viewed by others
Introduction
Rutaceae are represented in Madagascar by eight genera: Cedrelopsis Baill., Chloroxylon Scop., Citrus L., Fagaropsis Mildbr. ex Siebenl., Ivodea Capuron, Melicope J.R.Forst. & G.Forst., Vepris Comm. ex A.Juss., and Zanthoxylum L. (including Toddalia Juss.; Appelhans et al. 2018a). Of these, Citrus is not native, while Cedrelopsis is endemic to Madagascar and Ivodea is endemic to Madagascar and the Comoro Islands (Schatz 2001; Rabarimanarivo et al. 2015).
Ivodea is the only Malagasy genus that has never been included in a molecular phylogenetic study, and consequently, its phylogenetic position and biogeographic affinities are unknown. The genus has recently been revised, and the number of published species has risen from 17 to 30 (Rabarimanarivo et al. 2015). Ivodea was believed to be endemic to Madagascar until Labat et al. (2005) described a species from the Comoro Islands. Of the 30 currently accepted species, 27 are endemic to Madagascar, two are endemic to the Comoro Islands, and one species occurs on both Madagascar and the Comoro Islands. Ivodea is morphologically characterized by a membranaceous and brittle testa, seeds without endosperm, as well as the lack of a nectary disc (Capuron 1961; Kubitzki et al. 2011; Rabarimanarivo et al. 2015). Euodia J.R.Forst. & G.Forst. and Melicope have been hypothesized to be the closest relatives of Ivodea (Capuron 1961; Rabarimanarivo et al. 2015). Euodia resembles Ivodea in its thin and brittle seed coat, but Euodia is restricted to a relatively small area in Australasia and the West Pacific (Hartley 2001; Kubitzki et al. 2011; Appelhans et al. 2014, 2018b). A close relationship of the two genera would therefore be surprising from a biogeographic perspective, although it is not unconceivable since many examples of largely disjunct sister groups at various taxonomic levels exist (e.g. Le Roux et al. 2014). Melicope has a broad distribution throughout southern, eastern and SE Asia, Malesia, Australasia and many Pacific Islands as far east as the Hawaiian and Marquesas Islands. Additionally, Melicope is also known from Madagascar and the Mascarene Islands (Hartley 2001). From a biogeographic point of view, a close relationship of Ivodea and Melicope would not be surprising. Yet, the membranaceous and brittle testa, and the absence of endosperm and nectary disc in Ivodea are not indicative of a close affinity with Melicope (Perrier de la Bâthie 1950; Hartley 2001).
Apart from Euodia/Melicope, Ivodea has not yet been compared with other Rutaceae genera. In order to place Ivodea in the Rutaceae phylogeny and to evaluate whether Euodia and Melicope are indeed among the closest relatives of Ivodea, we assembled a dataset that contains representatives of all major clades of Rutaceae to which we added newly generated sequence data for Ivodea. Taxon sampling was based on previous phylogenetic studies (Chase et al. 1999; Groppo et al. 2008, 2012; Morton and Telmer 2014) and includes all four currently recognized subfamilies (sensu Morton and Telmer 2014), 11 out of 13 informal taxonomic units (‘alliances’; all except the Neotropical Amyris and Polyaster alliances that contain two and seven genera, respectively) used by Kubitzki et al. (2011), and covers the distribution of the family.
The main goals of the present study are to: (1) determine the phylogenetic position of Ivodea and (2) identify its biogeographic affinity. All Rutaceae genera from Madagascar except Ivodea have been sampled in previous phylogenetic studies. With the last previously unsampled genus included, an additional goal of this study is to (3) evaluate the biogeographic affinities of Malagasy Rutaceae as a whole.
Materials and methods
Taxon sampling
Our final dataset includes 94 samples representing 63 of the 154 genera of Rutaceae (Kubitzki et al. 2011) and contains samples of all major Rutaceae clades and all genera occurring on Madagascar (Schatz 2001; Groppo et al. 2008). Subfamily Aurantioideae, the diverse South American Galipeinae and the Southern African Diosmeae are represented only by few samples since they are not closely related to any Malagasy taxon, and initial BLAST searches and phylogenetic tree reconstructions using our newly generated data revealed no close relationship between Ivodea and these three groups. Five genera (out of eight) of subfamily Cneoroideae, which has been shown to be the sister group to Rutaceae s.s. (Appelhans et al. 2011), are included as outgroups.
Samples throughout the distributional range have been included for all genera occurring in Madagascar except Chloroxylon and Vepris. Chloroxylon has a disjunct distribution in Madagascar (two species), southern India and Sri Lanka (one species; Kubitzki et al. 2011), and only sequence data for the species from India and Sri Lanka were available. Vepris is represented by 13 out of about 80 species (Kubitzki et al. 2011) because BLAST searches revealed that Vepris species had the highest sequence similarity compared to our newly generated data for Ivodea. All GenBank entries for Vepris represent mainland African species, so we also sequenced a Vepris specimen from Madagascar. However, the genus also occurs on the Mascarenes, as well as in tropical Arabia and SW India, so that our sampling does not cover the whole distribution range. Several samples have been included for the species-rich and widespread genera Melicope and Zanthoxylum (Appelhans et al. 2014, 2018a).
Molecular laboratory work
DNA was extracted from herbarium samples using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. We initially amplified the internal transcribed spacer (ITS) region for the Ivodea samples using primers ITS4 and ITS5 (White et al. 1990) and performing a PCR with 35 cycles of 1 min at 95 °C, 1 min at 52 °C and 1 min at 72 °C, including initial denaturation of 7 min at 95 °C and final elongation steps of 7 min at 72 °C. PCR products were cleaned using ExoSAP-IT (affymetrix USB, Cleveland, Ohio, USA) and sequenced on an ABI 3100 capillary sequencer at Microsynth Seqlab (Göttingen, Germany). Forward and reverse sequences were combined, and the consensus sequence manually edited using the CLC Genomics Workbench 11.0.1 (QIAGEN, Aarhus, Denmark) and BLASTed against the nucleotide database at GenBank. The best matches belonged to the genus Vepris, and we decided to assemble a dataset consisting of ITS and trnL–trnF for our analyses since this marker set was used in the only published phylogenetic study of Vepris (Morton 2017). The trnL–trnF region was amplified and sequenced in two pieces using primers c, d, e and f (Taberlet et al. 1991) and applying the same PCR and sequencing protocols as outlined for ITS.
No ITS sequences could be obtained from GenBank for six genera. We had material at our disposal for three of them (Cedrelopsis, Ptaeroxylon and Spathelia) and complemented the data for these genera. Voucher information and GenBank numbers for all accessions are presented in Tables 1 and 2.
Alignment and phylogenetic analyses
Forward and reverse sequences were merged and edited manually using the CLC Genomics Workbench. Sequence alignments were generated using MUSCLE (Katoh et al. 2002; Katoh and Toh 2008) and edited by hand in Mesquite 3.40 (Maddison and Maddison 2015). The trnL–trnF alignment had a length of 1280 bp and contained 540 variable sites (42.2%) of which 286 (22.3%) were parsimony informative. The length of the ITS alignment was 743 bp of which 511 (68.8%) were variable and 379 (51.0%) were parsimony informative. Three regions with a combined length of 31 bp in the trnL–trnF alignment and two regions with a combined length of 65 bp in the ITS alignment could not be aligned with confidence and were excluded from the analyses.
Phylogenetic analysis of the ITS and trnL–trnF datasets as well as the two datasets combined was carried out using Bayesian inference (BI) and maximum likelihood (ML). Analyses were performed on the high-performance computing (HPC) cluster of Göttingen University. MrBayes 3.2.6. (Ronquist et al. 2012) was used for BI. The best-fitting substitution models were determined using jModelTest 2.1.3 (Darriba et al. 2012) and were set to GTR + G and GTR + I + G for trnL–trnF and ITS, respectively. BI analyses consisted of two independent Markov chain Monte Carlo (MCMC) runs with four chains each, which were observed for 10 million generations, and a tree was sampled every 100th generation. All runs reached stationarity (standard deviation of split frequencies < 0.01) after the 10 million generations. After inspecting the results in Tracer 1.6.0 (Rambaut et al. 2014; all effective sampling size [ESS] values were above 200) and removing 25% of the trees as burn-in, 50% majority-rule consensus trees were calculated in MrBayes. Only branches with posterior probability (pp) values of ≥ 0.95 pp were considered as supported.
ML analyses were carried out using RAxML 8.2.4 (Stamatakis 2014). The substitution models were specified as in the BI analyses for the single-marker analyses. For the combined analyses of trnL–trnF and ITS, the dataset was partitioned and the GTR + G model was used. All ML searches consisted of 1000 rapid bootstrap analyses followed by a search for the best-scoring tree. Branches with bootstrap (bs) values of 50–69 were considered to have low support, branches with bs values of 70–89 were regarded to have moderate support, and branches with bs values ≥ 90 were regarded as strongly supported.
The alignments and all consensus trees have been deposited at TreeBASE (https://purl.org/phylo/treebase/phylows/study/TB2:S24384).
Results
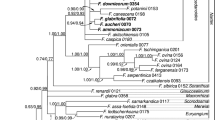
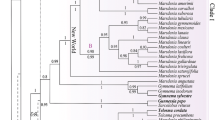
The consensus trees based on trnL–trnF (Fig. 1) and ITS (Fig. 2) alone showed congruent patterns of relationships, but differed in the resolution of clades. The more conserved trnL–trnF region provided better resolution at deeper nodes in the phylogeny, while ITS had better resolution towards the tips. There were supported conflicts between the trnL–trnF and ITS datasets concerning the position of Helietta Tul. (sister to Esenbeckia Brid. in the ITS dataset, but sister to Metrodorea A.St.-Hil. in the trnL–trnF dataset) and of several branches within Vepris. The concatenation might therefore be problematic, and we show the results for both the single-marker analyses and the combined analyses in the results and discussion, consequently.
Bayesian 50% majority-rule consensus tree based on the analysis of the ITS dataset. Posterior probabilities (pp; left) and the bootstrap values (bs; right) of the maximum likelihood analysis are displayed next to the branches. Asterisks indicate branches with high support ≥ 0.95 pp or ≥ 90% bs, respectively, and a hyphen represents unsupported branches in the ML analysis. Taxa native to Madagascar are highlighted in bold. Species epithets are included only for genera with several accessions, and numbers indicate several accessions of a species
Bayesian 50% majority-rule consensus tree based on the trnL–trnF dataset. Posterior probabilities (pp; left) and the bootstrap values (bs; right) of the maximum likelihood analysis are displayed next to the branches. Asterisks indicate branches with high support ≥ 0.95 pp or ≥ 90% bs, respectively, and a hyphen represents unsupported branches in the ML analysis. Taxa native to Madagascar are highlighted in bold. Species epithets are included only for genera with several accessions, and numbers indicate several accessions of a species
All consensus trees (trnL–trnF and ITS alone and the two makers combined; Figs. 1, 2, 3) reveal a close relationship between Ivodea and Vepris. While the two genera form a polytomy in the analyses based on the trnL–trnF dataset (1.00 pp, 100bs), the analyses based on ITS alone and trnL–trnF and ITS combined reveal a sister-group relationship of the two genera (ITS: 0.96 pp, 87bs; combined: 1.00 pp, 100bs) and support monophyly of both genera (Ivodea: ITS: 0.99 pp, 83bs; combined: 1.00 pp, 88bs; Vepris: ITS: 0.90 pp, 54bs; combined: 0.94 pp, 60bs). This study is the first to sample a Malagasy species of Vepris in a molecular phylogenetic context (V. elliottii (Radlk.) I.Verd.); it is here resolved as sister to the African representatives of Vepris albeit with only low support (ITS: 0.72 pp, < 50bs; combined: 0.92 pp, 55bs). Euodia and Melicope, which have been hypothesized to be the closest relatives of Ivodea, are only distantly related to it. Both Euodia and Melicope belong to a species-rich clade of Asian, Malesian and Australasian genera, which is sister to the Vepris–Ivodea clade (Figs. 1, 2, 3).
Bayesian 50% majority-rule consensus tree based on the analysis of the combined (trnL–trnF, ITS) dataset. Posterior probabilities (pp; left) and the bootstrap values (bs; right) of the maximum likelihood analysis are displayed next to the branches. Asterisks indicate branches with high support ≥ 0.95 pp and ≥ 90% bs, respectively, and a hyphen represents unsupported branches in the ML analysis. Taxa native to Madagascar are highlighted in bold. Species epithets are included only for genera with several accessions, and numbers indicate several accessions of a species
This is also the first time that more than one species of Fagaropsis is included in a phylogenetic analysis. Our results provide the first evidence for the monophyly of the genus (ITS: 0.99 pp, 87bs; trnL–trnF: 1.00 pp, 96bs; combined: 1.00 pp, 100 bs). Fagaropsis is confirmed to be part of the proto-Rutaceae clade (Appelhans et al. 2018a) and is sister to the Asian Phellodendron Rupr. and Tetradium Lour. (ITS: unresolved; trnL–trnF: 1.00 pp, 94bs; combined: 0.99 pp, 94bs). The relationships between the three Fagaropsis specimens are not resolved in the trnL–trnF dataset, but the ITS and the combined datasets reveal a grouping of the two sampled African specimens which are sister to the single sampled Malagasy specimen (ITS: 0.94 pp, 100bs; combined: 0.94 pp, 100bs).
Among the Rutaceae genera that contain species in Madagascar, only Ivodea and Vepris are close relatives (sister genera). All other genera with Malagasy representatives are not closely related to one another. Malagasy Rutaceae have representatives belonging to three of the four currently recognized subfamilies (all except Aurantioideae), with Cedrelopsis in Cneoroideae, Chloroxylon in Rutoideae and all others in the largest subfamily, Amyridoideae (sensu Morton and Telmer 2014).
Discussion
Phylogenetic placement of Ivodea
Our results reveal a sister-group relationship between the genera Ivodea and Vepris. Ivodea has been linked to Euodia and Melicope in previous studies (Capuron 1961; Rabarimanarivo et al. 2015). Euodia has been reduced from about 120 to seven species restricted to New Guinea, Australia and Pacific archipelagos as far east as Vanuatu (naturalized in other South Pacific Islands), and most of the species that were previously placed in Euodia have been transferred to Melicope and Tetradium (Hartley 1981, 2001). Species with a thin and brittle testa belong to Euodia s.s., which forms a clade with Brombya F.Muell., Perryodendron T.G.Hartley and Pitaviaster T.G.Hartley (Appelhans et al. 2014; Figs. 1, 2, 3). These three genera are also characterized by a thin and brittle testa, and they have overlapping distribution ranges in the Moluccas, New Guinea, tropical Australia and the western part of the South Pacific as far east as Vanuatu (Kubitzki et al. 2011; Appelhans et al. 2014). Moreover, the brittle seed coat and the lack of a nutritious and spongy exotesta probably make Euodia and its allies unsuited for long-distance dispersal via endozoochory (Appelhans et al. 2014). It would have been rather surprising if Ivodea were closely related to Euodia and its relatives given the large distance to Madagascar and no apparent suitability for bird dispersal (Appelhans et al. 2014). However, there are examples of Asian–Malagasy disjunctions of plants that lack obvious adaptations for long-distance dispersal. A well-known example is Nepenthes (Meimberg et al. 2001; Biswal et al. 2018). Moreover, long-distance dispersal events might not have been caused by the standard dispersal vector(s), but either by unusual behaviour of the standard vector or by a different vector (Higgins et al. 2003; Nathan et al. 2008). A close relationship of Ivodea and Melicope would have been more conceivable from a biogeographic point of view. Melicope has a broad distribution throughout southern, eastern and SE Asia, Malesia, Australasia and many Pacific Islands as far east as the Hawaiian and Marquesas Islands. Additionally, the genus is known from Madagascar and the Mascarene Islands (Hartley 2001). Melicope has a broad morphological circumscription (e.g. fruits follicular or capsular, monoecious or dioecious flowers), but species in some clades within Melicope and especially the Malagasy species resemble Ivodea in most morphological characters. Yet, the two genera differ substantially in the anatomy of the testa (membranaceous and brittle in Ivodea vs. thick sclerotesta and nutritious sarcotesta in Melicope), and the absence of endosperm and a nectary disc (Ivodea) vs. copious endosperm and a well-developed disc (Melicope) (Perrier de la Bâthie 1950; Hartley 2001).
The relationship of Ivodea and Vepris was surprising at first glance because of their different fruit types. On the other hand, Rabarimanarivo et al. (2015) mentioned that some species of Ivodea closely resemble the unifoliolate species of Vepris vegetatively as well as in flowering specimens. Engler (1931) based his Rutaceae classification mainly on fruit characters and placed Vepris in the subfamily Toddalioideae (Toddalieae–Toddaliinae) together with genera with exclusively drupaceous fruits. Engler’s treatment predated the recognition of Ivodea, but Capuron (1961) placed Ivodea in subfamily Rutoideae (Xanthoxyleae–Evodiinae), defined by the presence of dehiscent fruits. The separation into subfamilies with dehiscent and non-dehiscent fruits seems straightforward, but Engler (1931) made an exception by including the monotypic Chilean genus Pitavia Molina in Rutoideae despite its having drupaceous fruits. Since then, phytochemical and molecular phylogenetic studies have revealed that the two subfamilies are largely polyphyletic and that fruit characters are not good indicators of relationships at deeper phylogenetic scales in Rutaceae (Poon et al. 2007; Waterman 2007; Groppo et al. 2008). Several examples of pairs of taxa have been identified which have dehiscent and non-dehiscent fruits, respectively, or in which a taxon with one of these fruit types is nested within a clade that otherwise contains members with the other fruit type. These examples include Melicope (dehiscent) and Acronychia J.R.Forst. & G.Forst. (indehiscent); Sarcomelicope Engl./Dutaillyea Baill./Comptonella Baker f./Picrella Baill. (all indehiscent) and Melicope p.p. (dehiscent); Euodia (dehiscent) and Pitaviaster (indehiscent) (Appelhans et al. 2014); Zanthoxylum (dehiscent) and Toddalia (indehiscent; now merged into Zanthoxylum); Phellodendron (indehiscent) and Tetradium (dehiscent) (Appelhans et al. 2018a); as well as Acradenia Kippist (dehiscent) and Crossosperma T.G.Hartley (indehiscent) (Bayly et al. 2013). Ivodea and Vepris represent one more example in this growing list.
A characteristic feature of Ivodea is the lack of endosperm (Capuron 1961; Rabarimanarivo et al. 2015). Seeds lacking endosperm are typical for the southern African Diosma L. alliance and Aurantioideae, but are rare in other Rutaceae (Kubitzki et al. 2011). Interestingly, Vepris is polymorphic in this respect, and species may have copious or sparse endosperm or it may be completely lacking (Kubitzki et al. 2011). Moreover, Ivodea and Vepris share a characteristic thin seed coat (Kubitzki et al. 2011). Phyllotaxis is important for recognizing genera and major clades in Rutaceae (e.g. Appelhans et al. 2014); however, both Ivodea and Vepris exhibit variation in phyllotaxis, from alternate to subopposite or opposite (Ivodea; Rabarimanarivo et al. 2015) and alternate to rarely subopposite (Vepris; Schatz 2001). For most other morphological characters, Ivodea represents a subset of the character states of the more broadly defined Vepris, e.g. dioecious (in Ivodea) vs. dioecious or polygamous (in Vepris), leaves unifoliolate versus 1-, 3–9(-12)-foliolate, flowers 4(-5)-merous versus (3-)4(-5)-merous and 2 versus 1 or 2 ovules per locule. The only entirely discriminating characters between the two genera are the lack of a nectary disc in Ivodea and the different fruit types (Kubitzki et al. 2011).
Biogeographic affinities of Malagasy Rutaceae
The highest proportion of Malagasy angiosperm colonists stems from Africa (Yoder and Nowak 2006). However, there is also a high proportion of Malagasy lineages with an Indian, E Asian, SE Asian and Australian origin (Yoder and Nowak 2006; Buerki et al. 2013; Janssens et al. 2016). The direction of dispersal has not always been from these areas to Madagascar, but Madagascar has also been identified as the source of dispersal events (Bacon et al. 2016). The Asian affinities of the Malagasy flora have been attributed to vicariance events resulting from the breakup of Gondwana by some authors, but most time-calibrated molecular phylogenetic studies have revealed a much more recent colonization of Madagascar (Warren et al. 2010; Buerki et al. 2013; Bacon et al. 2016). Instead of Gondwanan vicariance, long-distance dispersal and stepping-stone dispersal have been proposed as the major mechanisms of migrations from Asia to Madagascar. A prominent route for stepping-stone dispersal might have been from Southern India via several Indian Ocean archipelagos to Madagascar and the Mascarenes during times of lower sea levels (Schatz 1996; Warren et al. 2010).
Now that DNA sequence data are available from representatives of all currently recognized genera of Malagasy Rutaceae, a first comparison of origins of Malagasy Rutaceae can be made. The sister-taxon relationships of Malagasy Rutaceae suggest African and Asian affinities, although limited taxon sampling and relatively old and isolated clades prevent a conclusive inference in some cases (Table 3). This is particularly true for Chloroxylon, for which only one of its three species could be sampled and only part of the distribution is covered, and Vepris, whose immediate relatives are not known and for which only a part of the distribution likewise is covered in our sampling.
The closest relatives of Cedrelopsis (eight species) are the monotypic S African genus Ptaeroxylon Eckl. & Zeyh. and the monotypic E African genus Bottegoa Chiov. The former might either be sister to Cedrelopsis (Razafimandimbison et al. 2010) or nested within it, although this topology has very low support (Appelhans et al. 2011). An Ancestral Area Reconstruction suggested an African origin for the three genera (Appelhans et al. 2012). Since only the sister-group relationship of Cedrelopsis and Ptaeroxylon had strong support in previous analyses, an African origin for Cedrelopsis is likely. If future studies provide evidence for Ptaeroxylon being nested within Cedrelopsis, a Malagasy origin of the two genera and a subsequent colonization to mainland Africa by the ancestor of Ptaeroxylon would be the most parsimonious scenario. The split of Cedrelopsis/Ptaeroxylon from Bottegoa and the initial diversification of Cedrelopsis/Ptaeroxylon have been dated to the Oligocene and Miocene (Appelhans et al. 2012), so a long-distance dispersal event from mainland Africa (or from Madagascar in case the alternative topology will be supported in future analyses) is likely.
A single exemplar of Chloroxylon has been included in several deeper-level phylogenetic analyses (Chase et al. 1999; Groppo et al. 2008; Morton and Telmer 2014; Appelhans et al. 2016), and it was not possible to sample additional specimens in the present study. Thus, the monophyly of this genus with a disjunct distribution in Madagascar, India and Sri Lanka remains untested using molecular methods. From a morphological perspective, the genus is characterized by some unusual features for Rutaceae (e.g. six ovules per locule, 3-valved capsule, apically winged seeds; Schatz 2001), so that morphology strongly suggests monophyly of the genus. Assuming that the genus is monophyletic, it remains to be tested whether the origin of Chloroxylon is in Madagascar with a dispersal to India and Sri Lanka, or vice versa. The closest relatives of Chloroxylon are the rare central Chinese Psilopeganum Hemsl., the Asian and Malesian Boenninghausenia Rchb., Thamnosma Torr. & Frém., which has a disjunct distribution in N America, Somalia and southern Africa, and the Macaronesian, Mediterranean to SW Asian Ruta L. (Kubitzki et al. 2011; Appelhans et al. 2016). Given the large and disjunct distribution of the clade and because monophyly of Chloroxylon has not yet been tested using molecular methods, the biogeographic origin of Chloroxylon remains unresolved.
Previous studies have only included a single species of Fagaropsis. The genus consists of four species, which occur in central and eastern Africa (two species) and on Madagascar (two species; Kubitzki et al. 2011). We sampled African and Malagasy species and provide the first evidence for monophyly of the genus. Fagaropsis belongs to a group of four genera referred to as proto-Rutaceae, which also includes the Asian Phellodendron and Tetradium, as well as the pantropical Zanthoxylum (Ling et al. 2009; Appelhans et al. 2018a). Despite the predominance of Asian taxa in this group, the oldest known fossils are European (Chandler 1961; Gregor 1989), so a Eurasian origin of the proto-Rutaceae has been proposed (Appelhans et al. 2018a). The ancestor of Fagaropsis likely originated in Eurasia and either colonized Africa first and later dispersed to Madagascar or vice versa.
The second member of proto-Rutaceae native to Madagascar is Zanthoxylum, and the Malagasy members of this large, pantropical genus belong to two separate lineages within it. Zanthoxlyum contains about 225 species and is divided into four main clades, and all African, Malagasy and Mascarene species form a clade that is sister to the remainder of the genus (taxa from Asia, the Americas and the Pacific; Appelhans et al. 2018a). The Malagasy and Mascarene endemic species are deeply nested within the African clade of Zanthoxylum (Appelhans et al. 2018a); thus, the phylogenetic reconstructions provide support for an African origin of the ancestor of the Malagasy and Mascarene species. The split of the Malagasy and Mascarene species from their African ancestors has been dated from the Oligocene to the Miocene (Appelhans et al. 2018a). Zanthoxylum asiaticum (formerly Toddalia asiatica) represents the second lineage of Zanthoxylum that occurs in Madagascar. This species has a large distribution, ranging from E Africa, the Comoros, Madagascar and the Mascarene Islands to S and SE Asia and the Philippines (Schatz 2001; Kubitzki et al. 2011). It was resolved as sister to the African clade of Zanthoxylum (Appelhans et al. 2018a), but in another study it was found to be sister to a main Asian clade (M.S. Appelhans, unpublished). An increased sampling of Z. asiaticum specimens throughout its distribution is needed in future phylogenetic analyses to determine whether this taxon originated in Africa and migrated to Asia, or vice versa, and which area is the source for the Malagasy populations.
The Malagasy and Mascarene species of Melicope have been resolved as monophyletic within Melicope section Lepta (Lour.) T.G.Hartley although several internal nodes had low support. They are deeply nested within this species-rich (about 234 species) and widespread genus (Kubitzki et al. 2011) and their closest relatives are taxa from mainland S and SE Asia as well as from Borneo (Appelhans et al. 2014). The split of the Malagasy and Mascarene lineage from their closest relatives has been dated to the Pliocene (Appelhans et al. 2014, 2018b). While Appelhans et al. (2014, 2018b) did not sample a crucial species from India and taxon sampling from Sumatra was low, an Asian origin for the Malagasy and Mascarene species is evident, but further studies are needed to evaluate whether the source area was India, SE Asia or Malesia and whether a single long-distance dispersal event or a stepping-stone dispersal via Indian Ocean archipelagos led to the presence of Melicope in Madagascar and the Mascarene Islands.
The sister position of Ivodea to Vepris, with the Malagasy V. elliotii resolved as sister to the African Vepris species, suggests the importance of Madagascar in the origin and evolution of the Ivodea–Vepris clade. The two earliest diverged clades within the Ivodea–Vepris clade from Madagascar (Figs. 1, 2, 3) suggest an origin of the group in Madagascar and one migration event to Africa, which led to the species-rich African clade of Vepris. A less parsimonious alternative hypothesis is an African origin of the Ivodea–Vepris clade followed by two independent migration events from Africa to Madagascar. A denser sampling of Vepris from Madagascar and the Mascarenes, and an inclusion of the two species from Arabia and India, are needed for a conclusive assessment. The African–Malagasy–Indian (S Asian) disjunct distribution has been reported in other plant lineages (Raven and Axelrod 1974), e.g. in Cyphostemma (Vitaceae, Hearn et al. 2018; Wen et al. 2018) and Exacum (Gentianaceae, Yuan et al. 2005; Pirie et al. 2015). The closest relatives of the Ivodea–Vepris lineage (Figs. 1, 2, 3; Groppo et al. 2008) might be a large SE Asian, Malesian and Australasian clade consisting of the Boronia Sm. alliance, the Bosistoa F.Muell. alliance and the Euodia alliance (sensu Kubitzki et al. 2011). Given these relationships, it is conceivable that the ancestor of Vepris and Ivodea might have dispersed from SE Asia, Malesia or Australasia to Madagascar and Africa. Alternatively, the species-rich SE Asian, Malesian and Australasian clade might have its origin in Madagascar or Africa. A well-supported backbone phylogeny is needed to elucidate the directionality of this dispersal event. The split of Vepris from the Boronia–Bosistoa–Euodia group has been dated to the Late Paleocene to Early Eocene (Appelhans et al. 2012). During these epochs, the migration of the Indian tectonic plate, and islands such as the Seychelles, which presumably existed as stepping stones (Schatz 1996; Yoder and Nowak 2006; Warren et al. 2010; Scotese 2014), might have facilitated dispersal. Further research with enhanced taxon sampling of the Malagasy Vepris species is needed in order to infer the directionality of the dispersal events. If our hypothesis about the SE Asian, Malesian or Australasian origin of the Ivodea–Vepris clade proves to be correct, the original hypothesis of a relationship of Ivodea with SE Asian taxa would be confirmed even though the closest relatives are not Euodia or Melicope (Capuron 1961).
Conclusions
Our analyses suggest a sister-group relationship of Ivodea and Vepris. A close relationship of Ivodea with the largely Asian and Pacific genera Euodia and Melicope, as was suggested in previous, morphology-based studies (Capuron 1961; Rabarimanarivo et al. 2015), is not supported in our study. Based on our sampling, the two early-diverged lineages within the Ivodea–Vepris clade represent Malagasy species, while the remainder of the clade consists of African species. The two sister clades of Ivodea–Vepris consist of Asian taxa or taxa with an Asian origin, although the support for these relationships is moderate to low. These results suggest that the Ivodea–Vepris clade may have originated in Asia and initially colonized Madagascar, followed by dispersal of Vepris to Africa. Species of Vepris from Arabia and India (one species each; Kubitzki et al. 2011) could not be sampled in this study. Future studies including these species will shed more light on the origin of this clade. Of the other Malagasy genera of Rutaceae, Cedrelopsis and one of the two Malagasy lineages of Zanthoxlyum are inferred to have an African origin, while Melicope originated in Asia. Fagaropsis has an African affinity, but it remains unclear whether the ancestor of the genus colonized Madagascar first (likely from Eurasia) followed by dispersal to Africa or whether Africa was colonized first. Further studies are needed to assess the monophyly and geographic origin of Chloroxylon and Zanthoxylum asiaticum. Both taxa have a large and disjunct distribution, and sampling throughout their distribution is needed for deeper insights into their evolutionary diversification.
References
Appelhans MS, Smets E, Razafimandimbison SG, Haevermans T, van Marle EJ, Rabarison H, Couloux A, Randrianarivelojosia M, Keßler PJA (2011) Phylogeny, evolutionary trends, and classification of the Spathelia/Ptaeroxylon clade: morphological and molecular insights. Ann Bot (Oxford) 107:1259–1277. https://doi.org/10.1093/aob/mcr076
Appelhans MS, Keßler PJA, Smets E, Razafimandimbison SG, Janssens SB (2012) Age and historical biogeography of the pantropically distributed Spathelioideae (Rutaceae, Sapindales). J Biogeogr 39:1235–1250. https://doi.org/10.1111/j.1365-2699.2012.02686.x
Appelhans MS, Wen J, Wagner WL (2014) A molecular phylogeny of Acronychia, Euodia, Melicope and relatives (Rutaceae) reveals polyphyletic genera and key innovations for species richness. Molec Phylogen Evol 79:54–68. https://doi.org/10.1016/j.ympev.2014.06.014
Appelhans MS, Krohm S, Manafzadeh S, Wen J (2016) Phylogenetic placement of Psilopeganum, a rare monotypic genus of Rutaceae (the citrus family) endemic to China. J Syst Evol 54:535–544. https://doi.org/10.1111/jse.12208
Appelhans MS, Reichelt N, Groppo M, Paetzold C, Wen J (2018a) Phylogeny and biogeography of the pantropical genus Zanthoxylum and its closest relatives in the proto-Rutaceae group (Rutaceae). Molec Phylogen Evol 126:31–44. https://doi.org/10.1016/j.ympev.2018.04.013
Appelhans MS, Wen J, Duretto M, Crayn D, Wagner WL (2018b) Historical biogeography of Melicope (Rutaceae) and its close relatives with a special emphasis on Pacific dispersals. J Syst Evol 56:576–599. https://doi.org/10.1111/jse.12299
Bacon CD, Simmons MP, Archer RH, Zhao LC, Andriantiana J (2016) Biogeography of the Malagasy Celastraceae: Multiple independent origins followed by widespread dispersal of genera from Madagascar. Molec Phylogen Evol 94:365–382
Bayly MJ, Holmes GD, Forster PI, Cantrill DJ, Ladiges PY (2013) Major clades of Australasian Rutoideae (Rutaceae) based on rbcL and atpB sequences. PLoS ONE 8:e72493
Biswal DK, Debnath M, Konhar R, Yanthan S, Tandon P (2018) Phylogeny and biogeography of carnivorous plant family Nepenthaceae with reference to the Indian pitcher plant Nepenthes khasiana reveals an Indian subcontinent origin of Nepenthes colonization in South East Asia during the Miocene epoch. Frontiers Ecol Evol 6:108. https://doi.org/10.3389/fevo.2018.00108
Buerki S, Devey DS, Callmander MW, Phillipson PB, Forest F (2013) Spatio-temporal history of the endemic genera of Madagascar. Bot J Linn Soc 171:304–329
Capuron R (1961) Contribution à l’étude de la ore forestière de Madagascar. III. Sur quelques plantes ayant contribué au peuplement de Madagascar. A. Rutacées nouvelles. 4. Ivodea R. Capuron gen. nov. (Rutoideae–Xanthoxyleae–Evodiinae). Adansonia 1:73–82
Chandler MEJ (1961) The lower Tertiary floras of Southern England, I, Paleocene Floras, London Clay Flora (supplement). British Museum, London
Chase MW, Morton CM, Kallunki JA (1999) Phylogenetic relationships of Rutaceae: a cladistics analysis of the subfamilies using evidence from rbcL and atpB sequence variation. Amer J Bot 86:1191–1999
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Meth 9:772
Engler A (1931) Rutaceae. In: Engler A, Harms H (eds) Die natürlichen Pflanzenfamilien—Band 19a. Wilhelm Engelmann, Leipzig, pp 187–359
Gregor HJ (1989) Aspects of the fossil record and phylogeny of the family Rutaceae (Zanthoxyleae, Toddalioideae). Pl Syst Evol 161:251–265
Groppo M, Pirani JR, Salatino MLF, Blanco SR, Kallunki JA (2008) Phylogeny of Rutaceae based on two non-coding regions from cpDNA. Amer J Bot 95:985–1005. https://doi.org/10.3732/ajb.2007313
Groppo M, Kallunki JA, Pirani JR, Antonelli A (2012) Chilean Pitavia more closely related to Oceania and Old World Rutaceae than to Neotropical groups: evidence from two cpDNA non-coding regions, with a new subfamilial classification of the family. Phytokeys 19:9–29
Hartley TG (1981) A revision of the genus Tetradium (Rutaceae). Garden’s Bull Singapore 34:91–131
Hartley TG (2001) On the taxonomy and biogeography of Euodia and Melicope (Rutaceae). Allertonia 8:1–341
Hearn DJ, Evans M, Wolf B, McGinty M, Wen J (2018) Dispersal is associated with morphological innovation, but not increased diversification, in Cyphostemma (Vitaceae). J Syst Evol 56:340–359
Higgins SI, Nathan R, Cain ML (2003) Are long-distance dispersal events in plants usually caused by nonstandard means of dispersal? Ecology 84:1945–1956
Janssens SB, Groeninckx I, De Block PJ, Verstraete B, Smets EF (2016) Dispersing towards Madagascar: Biogeography and evolution of the Madagascan endemics of the Spermacoceae tribe (Rubiaceae). Molec Phylogen Evol 95:58–66
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinf 9:286–298
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Kubitzki K, Kallunki JA, Duretto M, Wilson PG (2011) Rutaceae. In: Kubitzki K (ed) The families and genera of vascular plants, vol. 10. Springer, Berlin, pp 276–356
Labat JN, Pignal M, Pascal O (2005) Deux espèces nouvelles et une combinaison nouvelle chez les Rutaceae de l’Archipel des Comores. Monogr Syst Bot Missouri Bot Gard 104:362–365
Le Roux JJ, Strasberg D, Rouget M, Morden CW, Koordom M, Richardson DM (2014) Relatedness defies biogeography: the tale of two island endemics (Acacia heterophylla and A. koa). New Phytol 204:230–242
Ling KH, Wang Y, Poon WS, Shaw PC, But PPH (2009) The relationship of Fagaropsis and Luvunga in Rutaceae. Taiwania 54:338–342
Maddison WP, Maddison DR (2015) Mesquite: a modular system for evolutionary analysis, version 3.40 [online]. Available at: https://mesquiteproject.org. Accessed 15 Apr 2018
Meimberg H, Wistuba A, Dittrich P, Heubl G (2001) Molecular phylogeny of Nepenthaceae based on cladistics analysis of plastid trnK intron sequence data. Pl Biol 3:164–175
Morton CM (2017) Phylogenetic relationships of Vepris (Rutaceae) inferred from chloroplast, nuclear, and morphological data. PLoS ONE 12:e0172708. https://doi.org/10.1371/journal.pone.0172708
Morton CM, Telmer C (2014) New subfamily classification for the Rutaceae. Ann Missouri Bot Gard 99:620–641
Nathan R, Schurr FM, Spiegel O, Steinitz O, Trakhtenbrot A, Tsoar A (2008) Mechanisms of long-distance seed dispersal. Trends Ecol Evol 23:638–647
Perrier de la Bâthie H (1950) Rutacées. In: Humbert H (ed) Flore de Madagascar et des Comores 104e familie. Firmin-Didot et Cie, Paris, pp 1–89
Pirie MD, Litsios G, Bellstedt DU, Salamin N, Kissling J (2015) Back to Gondwanaland: can ancient vicariance explain (some) Indian Ocean disjunct plant distributions? Biol Lett 11:20150086
Poon WS, Shaw PC, Simmons MP, But PPH (2007) Congruence of molecular, morphological, and biochemical profiles in Rutaceae: a cladistic analysis of the subfamilies Rutoideae and Toddalioideae. Syst Bot 32:837–846
Rabarimanarivo M, Rakotonirina N, Phillipson PB, Lowry PP II, Labat JN, Pignal M (2015) Révision du genre Ivodea Capuron (Rutaceae), endémique de Madagascar et de l'archipel des Comores. Adansonia, sér 3 37:63–102
Rambaut A, Suchard MA, Xie D, Drummond AJ (2014) Tracer v1.6. Available at: https://tree.bio.ed.ac.uk/software/tracer/. Accessed 15 Apr 2018
Raven PH, Axelrod DI (1974) Angiosperm biogeography and past continental movements. Ann Missouri Bot Gard 61:539–673
Razafimandimbison SG, Appelhans MS, Rabarison H, Haevermans T, Rakotondrafara A, Rakotonandrasana SR, Ratsimbason M, Labat J-N, Keßler PJA, Smets E, Cruaud C, Couloux A, Randrianarivelojosia M (2010) Implications of a molecular phylogenetic study of the Malagasy genus Cedrelopsis and its relatives (Ptaeroxylaceae). Molec Phylogen Evol 57:258–265. https://doi.org/10.1016/j.ympev.2010.06.023
Ronquist F, Teslenko M, van der Mark P, Ayres D, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Schatz G (1996) Malagasy/Indo-Australo-Malesian phytogeographic connections. In: Lourenço WR (ed) Biogéographie de Madagascar. ORSTOM, Paris, pp 73–83
Schatz G (2001) Generic Tree Flora of Madagascar. Royal Botanic Gardens. Kew & Missouri Botanical Garden, St. Louis
Scotese CR (2014) Atlas of Paleogene Paleogeographic Maps (Mollweide Projection), Maps 8-15, vol. 1. The Cenozoic, PALEOMAP Atlas for ArcGIS. PALEOMAP Project, Evanston
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Pl Molec Biol 17:1105–1109
Warren BH, Strasberg D, Bruggemann JH, Prys-Jones RP, Thébaud C (2010) Why does the biota of the Madagascar region have such a strong Asiatic flavour? Cladistics 26:526–538. https://doi.org/10.1111/j.1096-0031.2009.00300.x
Waterman PG (2007) The current status of chemical systematics. Phytochemistry 68:2896–2903
Wen J, Lu LM, Nie ZL, Liu XQ, Zhang N, Ickert-Bond SM, Gerrath J, Manchester SR, Boggan J, Chen ZD (2018) A new phylogenetic tribal classification of the grape family (Vitaceae). J Syst Evol 56:262–272
White TJ, Burns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and amplifications. Academic Press, San Diego, pp 315–322
Yoder AD, Nowak MD (2006) Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annual Rev Ecol Evol Syst 37:405–431
Yuan Y-M, Wohlhauser S, Moller M, Klackenberg J, Callmander M, Kupfer P (2005) Phylogeny and biogeography of Exacum (Gentianaceae): a disjunctive distribution in the Indian Ocean Basin resulted from long distance dispersal and extensive radiation. Syst Biol 54:21–34
Acknowledgements
Open Access funding provided by Projekt DEAL. We are thankful to the curators of the Missouri Botanical Garden herbarium (MO) for providing leaf material for DNA analyses and to the colleagues from Naturalis (Leiden, the Netherlands) for sending DNAs used in an earlier publication (Appelhans et al. 2011). We also thank an anonymous reviewer for the very helpful and constructive comments that greatly improved this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights statement
No human participants or animals were involved in this research.
Additional information
Handling Editor: Alexander N. Sennikov.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Appelhans, M.S., Wen, J. Phylogenetic placement of Ivodea and biogeographic affinities of Malagasy Rutaceae. Plant Syst Evol 306, 7 (2020). https://doi.org/10.1007/s00606-020-01633-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00606-020-01633-3