Abstract
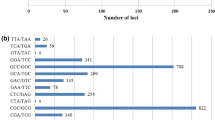
The genus Hibiscus comprises approximately 300 diverse species whose genetic relationships have not been extensively clarified. In this study, we evaluated the cross-species transferability of 102 expressed sequence tag-simple sequence repeat (EST-SSR) markers derived from the transcriptome of kenaf H. cannabinus L. and used them to assess the genetic diversity and inter-relationships of 87 accessions and 7 cultivars of 18 Hibiscus species. Among 102 markers, 100 (98%) were polymorphic in these 94 accessions/cultivars. The rates of cross-species transferability of the EST-SSR markers ranged from 83 H. trionum L. to 99% H. ponticus Rupr., with an average of 89.9%. A total of 871 alleles were detected using the 100 polymorphic EST-SSR markers; the number of alleles ranged from 2 (RBRC_Hc_ES_76) to 16 (RBRC_Hc_ES_80) and averaged 8.71. Polymorphism information content values varied from 0.15 (RBRC_Hc_ES_74) to 0.84 (RBRC_Hc_ES_3, RBRC_Hc_ES_95), with an average of 0.56. Phylogenetic and population structure analyses classified the 94 accessions/cultivars into two and two or three groups, respectively. Overall, our results demonstrate that the EST-SSR markers derived from the kenaf transcriptome were applied successfully to other Hibiscus species to provide useful information on the genetic diversity of this genus.


Similar content being viewed by others
References
Agarwal M, Shrivastava N, Padh H (2008) Advances in molecular marker techniques and their application in plant sciences. Plant Cell Rep 27:617–631
Akil HM, Omar MF, Mazuki AAM, Safiee S, Ishak ZAM, Bakar AA (2011) Kenaf fiber reinforced composite: a review. Mater Des 32:4107–4121
Anderson NO (2006) Flower breeding and genetics: issues, challenges and opportunities for the 21st century. Springer, Dordrecht, pp 479–489
Ayanbamiji TA, Ogundipe OT, Olowokudejo JD (2012) Taxonomic significance of the epicalyx in the genus Hibiscus (Malvaceae). Phytol Balc 18:135–140
Barbara T, Palma-Silva C, Paggi GM, Bered F, Fay MF, Lexer C (2007) Cross-species transfer of nuclear microsatellite markers: potential and limitations. Mol Ecol 16:3759–3767
Barik S, Senapati SK, Aparajita S, Mohapatra A, Rout GR (2006) Identification and genetic variation among Hibiscus species (Malvaceae) using RAPD marker. Z Naturfsch C 1:123–128
Boccacci P, Beltramo C, Sandoval Prando MA, Lembo A, Sartor C, Mehlenbacher SA, Botta R, Torello Marinoni D (2015) In silico mining, characterization and cross-species transferability of EST-SSR markers for European hazelnut (Corylus avellana L.). Mol Breed 35:21
Bragila L, Bruna S, Lanteri S, Mercuri A, Portis E (2010) An AFLP-based assessment of the genetic diversity within Hibiscus rosa-sinensis and its place within the Hibiscus genus complex. Sci Hortic 123:372–378
Bruna S, Portis E, Braglia L, Benedetti LD, Comino C, Acquadro A, Mercuri A (2009) Isolation and characterization of microsatellite markers from Hibiscus rosa-sinensis (Malvaceae) and cross-species amplifications. Conserv Genet 10:771–774
Calısır S, Ozcan M, Hacıseferogulları H, Yıldız MU (2005) A study on some physic-chemical properties of Turkey okra (Hibiscus esculenta L.) seeds. J Food Eng 68:73–78
Chen H, Chen H, Hu L, Wang L, Wang S, Wang ML, Cheng X (2017) Genetic diversity and population structure analysis of accessions in the Chinese cowpea [Vigna unguiculata (L.) Walp.] germplasm collection. Crop J 5:363–372
Cheng Z, Lu B-R, Sameshima K, Fu D-X, Chen J-K (2004) Identification and genetic relationships of kenaf (Hibiscus cannabinus L.) germplasm revealed by AFLP analysis. Genet Resour Crop Evol 51:393–401
Choudhary S, Sethy NK, Shokeen B, Bhatia S (2009) Development of chickpea EST-SSR markers and analysis of allelic variation across related species. Theor Appl Genet 118:591–608
Decroocq V, Fave MG, Hagen L, Bordenave L, Decroocq S (2003) Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor Appl Genet 106:912–922
Ding Q, Li J, Wang F, Zhang Y, Li H, Zhang J, Gao J (2015) Characterization and development of EST-SSRs by deep transcriptome sequencing in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Int J Genomics 2015:473028
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132
Feng SP, Li WG, Huang HS, Wang JY, Wu YT (2009) Development, characterization and cross-species/genera transferability of EST-SSR markers for rubber tree (Hevea brasiliensis). Mol Breed 23:85–97
Fryxell PA (1997) The American genera of Malvaceae—II. Brittonia 49:204–269
Gasic K, Han Y, Kertbundit S, Shulaev V, Iezzoni AF, Stover EW, Bell RL, Wisniewski ME, Korban SS (2009) Characteristics and transferability of new apple EST-derived SSRs to other Rosaceae species. Mol Breed 23:397–411
Guo W, Wang W, Zhou B, Zhang T (2006) Cross-species transferability of G. arboreum derived EST-SSRs in the diploid species of Gossypium. Theor Appl Genet 112:1573–1581
Gupta PK, Rustgi S, Sharma S, Singh R, Kumar N, Balyan HS (2003) Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol Genet Genomics 270:315–323
Harmon M, Lane T, Staton M, Coggeshall MV, Best T, Chen C-C, Liang H, Zembower N, Drautz-Moses DI, Hwee YZ, Schuster SC, Schlarbaum SE, Carlson JE, Gailing O (2017) Development of novel genic microsatellite markers from transcriptome sequencing in sugar maple (Acer saccharum Marsh.). BMC Res Notes 10:369
Hendrick PW (2001) Conservation genetics: where are we now? Trends Ecol Evol 16:629–636
Holton TA, Christopher JT, McClure L, Harker N, Henry RJ (2002) Identification and mapping of polymorphic SSR markers from expressed gene sequences of barley and wheat. Mol Breed 9:63–71
Jung SW, Kwon S-J, Ryu J, Kim J-B, Ahn J-W, Kim SH, Jo YD, Choi H-I, Im SB, Kang S-Y (2017) Development of EST-SSR markers through de novo RNA sequencing and application for biomass productivity in kenaf (Hiviscus cannabinus L.). Genes Genom 39:1139–1156
Kachecheba JL (1972) The cytotaxonomy of some species of Hibiscus. Kew Bull 27:425–433
Kaldate R, Rana M, Sharma V, Hirakawa H, Kumar R, Singh G, Chahota RK, Isobe SN, Sharma TR (2017) Development of genome-wide SSR markers in horsegram and their use for genetic diversity and cross-transferability analysis. Mol Breed 37:103
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK (2011) Microsatellite markers: an overview of the recent progress in plants. Euphytica 177:309–334
Kantety RV, Matthws ML, Rota DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barely, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Kapepula PM, Ngombe NK, Tshibangu PT, Tsumbu C, Franck T, Mouiths-Mickalad A, Mumba D, Tshala-Katumbay D, Serteyn D, Tits M, Angenot L, Kalenda TDP, Frédérich M (2017) Comparison of metabolic profiles and bioactivities of the leaves of three edible Congolese Hibiscus species. Nat Prod Res 31:2885–2892
Kong Q, Xiang C, Yang J, Yu Z (2011) Genetic variations of Chinese melon landraces investigated with EST-SSR markers. Hort Environ Biotechnol 52:163–169
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lee G-A, Kwon S-J, Park Y-J, Lee M-C, Kim H-H, Lee J-S, Lee S-Y, Gwag J-G, Kim C-K, Ma K-H (2011) Cross-amplification of SSR markers developed from Allium sativum to other Allium species. Sci Hortic 128:401–407
Litt M, Luty JA (1989) A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Human Genet 44:397–401
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Liu S, Yanlin A, Li F, Li S, Liu L, Zhou Q, Zhao S, Wei C (2018) Genome-wide identification of simple sequence repeats and development of polymorphic SSR markers for genetic studies in tea plant (Camellia sinensis). Mol Breed 38:59
Ma K-H, Kim N-S, Lee G-A, Lee S-Y, Lee J-K, Yi JY, Park Y-J, Kim T-S, Gwag J-G, Kwon S-J (2009) Development of SSR markers from studies of diversity in the genus Fagopyrum. Theor Appl Genet 119:1247–1254
Maganha EG, Halmenschlager RC, Rosa RM, Henriques JAP, Ramos ALLP, Saffi J (2010) Pharmacological evidences for the extracts and secondary metabolites from plants of the genus Hibiscus. Food Chem 118:1–10
Mariotti R, Cultrera NGM, Mousavi S, Baglivo F, Rossi M, Albertini E, Alagna F, Carbone F, Perrotta G, Baldoni L (2016) Development, evaluation, and validiation of new EST-SSR markers in olive (Olea europanea L.). Tree Genet Genomes 12:120
Meyer L, Causse R, Pernin F, Scalone R, Bailly G, Chauvel B, Delye C, Corre VL (2017) New gSSR and EST-SSR markers reveal high genetic diversity in the invasive plant Ambrosia artemisiifolia L. and can be transferred to other invasive Ambrosia species. PLoS ONE 12:e0176197
Mondini L, Noorani A, Pagnotta MA (2009) Assessing plant genetic diversity by molecular tools. Diversity 1:19–35
Moore SS, Sargeant LL, King TJ, Mattick JS, Georges M, Hetzel DJS (1991) The conservation of dinucleotide microsatellites among mammalian genomes allows the use of heterologous PCR primer pairs in closely related species. Genomics 10:654–660
Mulato BM, Moller M, Zucchi MI, Quecini V, Pinheiro JB (2010) Genetic diversity in soybean germplasm identified by SSR and EST-SSR markers. Pesqui Agropecu Bras 45:76–283
Nicot N, Chiquet V, Gandon B, Amihat L, Legeai F, Leroy P, Bernard M, Sourdille P (2004) Study of simple sequence repeat (SSR) markers from wheat expressed sequence tags (ESTs). Theor Appl Genet 109:800–805
Ogundajo AL, Ogunwande IA, Bolarinwa TM, Joseph OR, Flamini G (2014) Essential oil of the leaves of Hibiscus surattensis L. from Nigeria. J Essent Oil Res 26:114–117
Olaleye MT (2007) Cytotoxicity and antibacterial activity of methanolic extract of Hibiscus sabdariffa. J Med Plant Res 1:9–13
Pandey S, Ansari WA, Pandey M, Singh B (2018) Genetic diversity of cucumber estimated by morpho-physiological and EST-SSR markers. Physiol Mol Biol Plants 24:135–146
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pfeil BE, Brubaker CL, Craven LA, Crisp MD (2002) Phylogeny of Hibiscus and the Tribe Hibisceae (Malvaceae) using chloroplast DNA sequences of ndhF and the rpl16 intron. Syst Bot 27:333–350
Pfeil BE, Brubaker CL, Craven LA, Crisp MD (2004) Paralogy and orthology in the Malvaceae rpb2 gene family: investigation of gene duplication in Hibiscus. Mol Biol Evol 21:1428–1437
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Pritchard JK, Stephen M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Puckhaber LS, Stipanovic RD, Bost GA (2002) Analyses for flavonoid aglycones in fresh and preserved Hibiscus flowers. Trends in new crops and new uses. ASHS Press, Alexandria, pp 556–563
Reddy MRK, Rathour R, Kumar N, Katoch P, Sharma TR (2010) Cross-genera legume SSR markers for analysis of genetic diversity in Lens species. Plant Breed 129:514–518
Roe SMJ (1961) A taxonomic study of the indigenous Hawaiian species of the genus Hibiscus (Malvaceae). Pac Sci 15:3–32
Rozen S, Skaletsky H (1999) Primer3 on the WWW for general users and for biologist programmers. In: Misener S, Krawetz S (eds) Bioinformatics methods and protocols. Humma Press, Totowa, pp 365–386
Satya P, Karan M, Sarkar D, Sinha MK (2012) Genome synteny and evolution of AABB allotetraploids in Hibiscus section Furcaria revealed by interspecies hybridization, ISSR and SSR markers. Plant Syst Evol 298:1257–1270
Schlotterer C (2000) Evolutionary dynamics of microsatellite DNA. Chromosoma 109:365–371
Shaheen N, Ajab M, Yasmin G, Hayat MQ (2009a) Diversity of foliar trichomes and their systematic relevance in the genus Hibiscus (Malvaceae). Int J Agric Biol 11:279–284
Shaheen N, Khan MA, Hayat MQ, Yasmin G (2009b) Pollen morphology of 14 species of Abutilon and Hibiscus of the family Malvaceae (sensu stricto). J Med Plant Res 3:921–929
Shiferaw E (2013) Development and cross-species amplification of grass pea EST-derived markers. Afr Crop Sci J 21:153–160
Singh RK, Jena SN, Khan S, Yadav S, Banarjee N, Raghuvanshi S, Bhardwaj V, Dattamajumder SK, Kapur R, Solomon S, Swapna M, Srivastava S, Tyagi AK (2013) Development, cross-species/genera transferability of novel EST-SSR markers and their utility in revealing population structure and genetic diversity in sugarcane. Gene 524:309–329
Skovsted A (1935) Chromosome numbers in the Malvaceae. I. J Genet 31:263–296
Takayama K, Kajita T, Murata J, Tateishi Y (2006) Isolation and characterization of microsatellites in the sea hibiscus (Hibiscus Tiliaceus L., Malvaceae) and related hibiscus species. Mol Ecol Notes 6:721–723
Thiel T, Michalek W, Varshne RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Van Huylenbroeck JM, De Riek J, De Loose M (2000) Genetic relationships among Hibiscus syriacus, Hibiscus sinosyriacus and Hibiscus paramutabilis revealed by AFLP, morphology and ploidy analysis. Genet Resour Crop Evol 47:335–343
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55
Vasudeva N, Sharma SK (2008) Biologically active compound from the genus Hibiscus. Pharm Biol 46:145–153
Wang Z, Yan H, Fu X, Li X, Gao H (2013) Development of simple sequence repeat markers and diversity analysis in alfalfa (Medicago sativa L.). Mol Biol Rep 40:3291
Wei W, Qi X, Wang L, Zhang Y, Hua W, Li D, Lv H, Zhang X (2011) Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genom 12:451
Wilson FD (1994) The genome biogeography of Hibiscus L. section Furcaria DC. Genet Resour Crop Evol 41:13–25
Wilson FD (1999) Revision of Hibiscus section Furcaria (Malvaceae) in Africa and Asia. Bull Nat Hist Mus Lond (Botany) 29:47–79
Wilson ACC, Massonnet B, Simon JC, Prunier-Leterme N, Dolatti L, Llewellyn KS, Figueroa CC, Ramirez CC, Blackman RL, Estoup A, Sunnucks P (2004) Cross-species amplification of microsatellite loci in aphids: assessment and application. Mol Ecol Notes 4:104–109
Yan Z, Wu F, Luo K, Zhao Y, Yan Q, Zhang Y, Wang Y, Zhang J (2017) Cross-species transferability of EST-SSR markers developed from the transcriptome of Melilotus and their application to population genetic research. Sci Rep 7:17959
Zalapa JE, Guevas H, Zhu H, Steffan S, Senalik D, Zeldin E, McCown B, Harbut R, Simon P (2012) Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sicences. Am J Bot 99:193–208
Zhai L, Xu L, Wang Y, Cheng H, Chen Y, Gong Y, Liu L (2014) Novel and useful genic-SSR from de novo transcriptome sequencing of radish (Raphanus sativus L.). Mol Breed 33:611–624
Zhou Q, Luo D, Ma L, Xie W, Wang Y, Wang Y, Liu Z (2016) Development and cross-species transferability of EST-SSR markers in Siberian wildrye (Elymus sibiricus L.) using Illumina sequencing. Sci Rep 6:20549
Acknowledgements
This work was supported by the Nuclear R&D program of KAERI and Radiation Technology R&D Program (NRF-2017M2A2A6A05018538) through the National Research Foundation of Korea funded by the Ministry of Science and ICT. We thank Margaret Biswas, Ph.D., from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10722_2019_817_MOESM1_ESM.pptx
Supplementary Fig. S1. EST-SSR patterns showing polymorphism and transferability of 18 Hibiscus species based on the RBRC_Hc_ES_4, RBRC_Hc_ES_20, and RBRC_Hc_ES_79 loci (PPTX 319 kb)
Rights and permissions
About this article
Cite this article
Kim, J.M., Lyu, J.I., Lee, MK. et al. Cross-species transferability of EST-SSR markers derived from the transcriptome of kenaf (Hibiscus cannabinus L.) and their application to genus Hibiscus. Genet Resour Crop Evol 66, 1543–1556 (2019). https://doi.org/10.1007/s10722-019-00817-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-019-00817-2