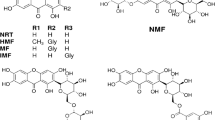
Abstract
Neurodegenerative disorders (NDDs) are associated with increased activities of the brain acetylcholinesterase (AChE), butyrylcholinesterase (BChE) and β-secretase enzyme (BACE1). Inhibition of these enzymes affords therapeutic option for managing NDDs such as Alzheimer’s disease (AD) and Parkinson’s disease (PD). Although, Gongronema latifolium Benth (GL) has been widely documented in ethnopharmacological and scientific reports for the management of NDDs, there is paucity of information on its underlying mechanism and neurotherapeutic constituents. Herein, 152 previously reported Gongronema latifolium derived-phytochemicals (GLDP) were screened against hAChE, hBChE and hBACE-1 using molecular docking, molecular dynamics (MD) simulations, free energy of binding calculations and cluster analysis. The result of the computational analysis identified silymarin, alpha-amyrin and teraxeron with the highest binding energies (-12.3, -11.2, -10.5 Kcal/mol) for hAChE, hBChE and hBACE-1 respectively as compared with those of the reference inhibitors (-12.3, -9.8 and − 9.4 for donepezil, propidium and aminoquinoline compound respectively). These best docked phytochemicals were found to be orientated in the hydrophobic gorge where they interacted with the choline-binding pocket in the A-site and P-site of the cholinesterase and subsites S1, S3, S3’ and flip (67–75) residues of the pocket of the BACE-1. The best docked phytochemicals complexed with the target proteins were stable in a 100 ns molecular dynamic simulation. The interactions with the catalytic residues were preserved during the simulation as observed from the MMGBSA decomposition and cluster analyses. The presence of these phytocompounds most notably silymarin, which demonstrated dual high binding tendencies to both cholinesterases, were identified as potential neurotherapeutics subject to further investigation.









Similar content being viewed by others
Data Availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Behl T, Makkar R, Sehgal A, Singh S, Sharma N, Zengin G et al (2021) Current Trends in Neurodegeneration: Cross Talks between oxidative stress, cell death, and inflammation. Int J Mol Sci 22:7432. https://doi.org/10.3390/ijms22147432
Dugger BN, Dickson DW (2017) Pathology of neurodegenerative diseases. Cold Spring Harb Perspect Biol 9:a028035
Feigin VL, Nichols E, Alam T, Bannick MS, Beghi E, Blake N et al (2019) Global, regional, and national burden of neurological disorders, 1990–2016: a systematic analysis for the global burden of Disease Study 2016. Lancet Neurol 18:459–480
Bozoki A, Giordani B, Heidebrink JL, Berent S, Foster NL (2001) Mild cognitive impairments predict dementia in nondemented elderly patients with memory loss. Archives of neurology. ;58:411 – 6.10.1001/archneur.58.3.411
Nichols E, Steinmetz JD, Vollset SE, Fukutaki K, Chalek J, Abd-Allah F et al (2022) Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: an analysis for the Global Burden of Disease Study 2019. The Lancet Public Health. ;7:e105-e25.10.1016/S2468-2667(21)00249-8
Hussain R, Zubair H, Pursell S, Shahab M (2018) Neurodegenerative diseases: regenerative mechanisms and novel therapeutic approaches. Brain Sci 8:177
Bar-Am O, Amit T, Kupershmidt L, Aluf Y, Mechlovich D, Kabha H et al (2015) Neuroprotective and neurorestorative activities of a novel iron chelator-brain selective monoamine oxidase-A/monoamine oxidase-B inhibitor in animal models of Parkinson’s disease and aging. Neurobiology of aging. ;36:1529 – 42.10.1016/j.neurobiolaging.2014.10.026
Hampel H, Mesulam M-M, Cuello AC, Farlow MR, Giacobini E, Grossberg GT et al (2018) The cholinergic system in the pathophysiology and treatment of Alzheimer’s disease. Brain 141:1917–1933
Atatreh N, Al Rawashdah S, Al Neyadi SS, Abuhamdah SM, Ghattas MA (2019) Discovery of new butyrylcholinesterase inhibitors via structure-based virtual screening. J Enzyme Inhib Med Chem 34:1373–1379
Marucci G, Buccioni M, Dal Ben D, Lambertucci C, Volpini R, Amenta F (2021) Efficacy of acetylcholinesterase inhibitors in Alzheimer’s disease. Neuropharmacology 190:108352
Klafki HW, Staufenbiel M, Kornhuber J, Wiltfang J (2006) Therapeutic approaches to Alzheimer’s disease. Brain: a journal of neurology. ;129:2840 – 55.10.1093/brain/awl280
Ravera E, Ciambellotti S, Cerofolini L, Martelli T, Kozyreva T, Bernacchioni C et al (2016) Solid-state NMR of PEGylated proteins. Angew Chem 128:2492–2495
Adewole KE, Gyebi GA, Ibrahim IM (2021) Amyloid β fibrils disruption by kolaviron: molecular docking and extended molecular dynamics simulation studies. Comput Biol Chem 94:107557
Brown MR, Radford SE, Hewitt EW (2020) Modulation of β-amyloid fibril formation in Alzheimer’s disease by microglia and infection. Front Mol Neurosci 13:609073
Hampel H, Lista S, Vanmechelen E, Zetterberg H, Giorgi FS, Galgani A et al (2020) β-Secretase1 biological markers for Alzheimer’s disease: state-of-art of validation and qualification. Alzheimer’s Research & Therapy. ;12:130.10.1186/s13195-020-00686-3
Ekong MB, Peter MD, Peter AI, Eluwa MA, Umoh IU, Igiri AO et al (2014) Cerebellar neurohistology and behavioural effects of Gongronema latifolium and Rauwolfia vomitoria in mice. Metab Brain Dis 29:521–527
Balogun M, Besong E, Obimma J, Mbamalu O, Djobissie S (2016) Gongronema latifolium: a phytochemical, nutritional and pharmacological review. J Physiol Pharmacol Adv 6:811–824
Ugochukwu N, Babady N, Cobourne M, Gasset S (2003) The effect ofGongronema latifolium extracts on serum lipid profile and oxidative stress in hepatocytes of diabetic rats. J Biosci 28:1–5
Nwanjo H, Okafor M, Oze G (2006) Anti-lipid peroxidative activity of Gongronema latifoluim in streptozotocin-induced diadetic rat. Nigerian J Physiological Sci. ;21
Adebajo A, Ayoola M, Odediran S, Aladesanmi A, Schmidt T, Verspohl E (2012) P 29: insulinotropic constituents and evaluation of ethnomedical claim of gongronema latifolium root and stem. Diabetes Metab 38:S115
Ibegbulem C, Chikezie P (2013) Hypoglycemic properties of ethanolic extracts of Gongronema latifolium, Aloe perryi, Viscum album and Allium sativum administered to alloxan-induced diabetic albino rats (Rattus norvegicus). Pharmacognosy Commun 3:12
Robert A, Luke U, Udosen E, Ufot S, Effiong A, Ekam V (2011) Anti-diabetic and Anti-hyperlipedemic Properties of Ethanolic Root Extract of Gongronema Latifolium (Utazi) on Streptozotocin (STZ) Induced Diabetic Rats.
Ogunyemi O, Gyebi A, Adebayo J, Oguntola J, Olaiya C (2020) Marsectohexol and other pregnane phytochemicals derived from Gongronema latifolium as α-amylase and α-glucosidase inhibitors: in vitro and molecular docking studies. SN Appl Sci 2:1–11
Ogunyemi O, Gyebi G, Saheed A, Paul J, Nwaneri-Chidozie V, Olorundare O et al Inhibition mechanism of alpha-amylase, a diabetes target, by a steroidal pregnane and pregnane glycosides derived from Gongronema latifolium Benth. Frontiers in molecular biosciences. 2022;9
Akpaso M, Orie N, Ebong P (2019) Administration of combined methanolic leaf extracts of vernonia amygdalina and gongronema latifolium enhanced glut 2 expression in the pancreas and downregulates serum caspase 3 activity of streptozotocin-induced diabetic wistar rats.
Ofoha PC, Nimenibo-Uadia RI Effect of Ethanolic Leaf Extract of Gongronema Latifolium on blood glucose and cholesterol levels in Alloxan-Induced Diabetic rats
Chukwudozie IK, Agbo MC, Ugwu KO, Ezeonu IM (2021) Oral administration of Gongronema lafifoliumLeaf Extract modulates gut microflora and blood glucose of Induced Diabetic rats. J Pure Appl Microbiol 15:346–355
Al-Hindi B, Yusoff NA, Ahmad M, Atangwho IJ, Asmawi MZ, Al-Mansoub MA et al (2019) Safety assessment of the ethanolic extract of Gongronema latifolium Benth. Leaves: a 90-day oral toxicity study in Sprague Dawley rats. BMC Complement Altern Med 19:1–10
SC O, Chinaka N (2013) Carbon tetrachloride induced renaltoxicity and the effect of aqueous extract of Gongronema latifolium in Wistar albino rats. Drug discovery 4:15–16
Imo C, Uhegbu FO, Glory IN (2015) Histological and Hepatoprotective Effect of Ethanolic Leaf Extract of Gongronema latifolium Benth in Acetaminophen-Induced hepatic toxicity in male albino rats. Int J Preventive Med Res. ;1
Johnkennedy N, Adamma E (2011) The protective role of Gongronema latifolium in acetaminophen induced hepatic toxicity in Wistar rats. Asian Pac J Trop Biomed 1:S151–S4
Adekanle E, Omozokpia UM (2015) Antioxidant potentials of Gongronema latifolium (utazi) leaf extracts. Biokemistri 27:85–88
Usoh I, Akpan H (2015) Antioxidative efficacy of combined leaves extracts of Gongronema latifolium and Ocimum gratissimum on streptozotocin induced diabetic rats. J Med Med Sci 2:88–95
Gyebi GA, Adebayo JO, Olorundare OE, Pardede A, Ninomiya M, Saheed AO et al (2018) Iloneoside: a cytotoxic ditigloylated pregnane glycoside from the leaves of Gongronema latifolium Benth. Nat Prod Res 32:2882–2886
Iweala EEJ (2015) Anti-cancer and free radical scavenging activity of some nigerian food plants in vitro. Int J cancer Res 11:41–51
Adebayo J, Ceravolo I, Gyebi G, Olorundare O, Babatunde A, Penna-Coutinho J et al (2022) Iloneoside, an antimalarial pregnane glycoside isolated from Gongronema latifolium leaf, potentiates the activity of chloroquine against multidrug resistant Plasmodium falciparum. Mol Biochem Parasitol 249:111474
Idowu ET, Ajaegbu HC, Omotayo AI, Aina OO, Otubanjo OA (2015) In vivo anti-plasmodial activities and toxic impacts of lime extract of a combination of Picralima nitida, Alstonia boonei and Gongronema latifolium in mice infected with chloroquine-sensitive Plasmodium berghei. Afr Health Sci 15:1262–1270
Ekong MB, Nwakanma AA (2017) Rauwolfia vomitoria and Gongronema latifolium extracts influences cerebellar cortex. Alzheimer’s. Dement Cogn Neurol 1:1–6
Akinnuga A, Bamidele O, Ekechi P, Adeniyi O (2011) Effects of an ethanolic leaf extract of Gongronema latifolium on haematological some parameters in rats. Afr J Biomedical Res 14:153–156
Owu D, Nwokocha C, Obembe A, Essien A, Ikpi D, Osim E (2012) Effect of Gongronema latifolium ethanol leaf extract on gastric acid secretion and cytoprotection in streptozotocin-induced diabetic rats. West Indian Med J. ;61
Egba S, Omeoga H, Njoku O (2014) Oral administration of methanol extract of Gongronema latifolium (utazi) Up-Regulates cytokine expression and influences the immune system in wistar albino rats. World Appl Sci J 31:745–750
Akani N, Nwachukwu C, Hakam I (2020) Evaluation of the antibacterial activity of Gongronema latifolium and Costus afer Leaf extracts on E. coli (ATCC 29455) and S. aureus (ATCC 25923). Int J Pathogen Res. :11–6
Enitan SS, Ehizibue OP, Adejumo EN, Akele YR, Owolabi TO (2017) Evaluation of the Antimicrobial potential of Gongronema latifolium extracts on some Wound-Associated Pathogens. Open Sci J Pharm Pharmacol 5:34–41
Ujong GO, Beshel JA, Nkanu E, Ubana OP, Ofem OE (2022) Ethanolic extract of Gongronema latifolium improves learning and memory in swiss albino mice. J Drug Delivery Ther 12:45–50
Nwanna EE, Oyeleye SI, Ogunsuyi OB, Oboh G, Boligon AA, Athayde ML (2016) In vitro neuroprotective properties of some commonly consumed green leafy vegetables in Southern Nigeria. NFS J 2:19–24. https://doi.org/10.1016/j.nfs.2015.12.002
Oyinloye BE, Iwaloye O, Ajiboye BO (2021) Polypharmacology of Gongronema latifolium leaf secondary metabolites against protein kinases implicated in Parkinson’s disease and Alzheimer’s disease. Sci Afr 12:e00826. https://doi.org/10.1016/j.sciaf.2021.e00826
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Imo C, Uhegbu FO (2015) Phytochemical analysis of Gongronema latifolium Benth leaf using gas chromatographic flame ionization detector. Int J Chem Biomol Sci 1:60–68
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: An open chemical toolbox. Journal of cheminformatics. ;3:33.10.1186/1758-2946-3-33
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. ;31:455 – 61.10.1002/jcc.21334
O’Boyle N, Banck M, James CA, Morley C, Vandermeersch T (2011) Hutchison GR Open Babel: an open chemical toolbox. J Cheminf 3:33
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B et al (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25
Bekker H, Berendsen H, Dijkstra E, Achterop S, Vondrumen R, Vanderspoel D et al (eds) (1993) Gromacs-a parallel computer for molecular-dynamics simulations. 4th international conference on computational physics (PC 92); : World Scientific Publishing
Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF (2004) A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem 25:1656–1676
Schüttelkopf AW, Van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr Sect D: Biol Crystallogr 60:1355–1363
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38
Valdés-Tresanco MS, Valdés-Tresanco ME, Valiente PA, Moreno E (2021) gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J Chem Theory Comput 17:6281–6291
Miller IIIBR, McGee TD Jr, Swails JM, Homeyer N, Gohlke H, Roitberg AE (2012) MMPBSA. py: an efficient program for end-state free energy calculations. J Chem Theory Comput 8:3314–3321
Xue W, Yang F, Wang P, Zheng G, Chen Y, Yao X et al (2018) What contributes to serotonin–norepinephrine reuptake inhibitors’ dual-targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem Neurosci 9:1128–1140
Tuccinardi T (2021) What is the current value of MM/PBSA and MM/GBSA methods in drug discovery? Taylor & Francis, pp 1233–1237
Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M (2015) PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res 43:W443–W7
Gyebi GA, Ogunyemi OM, Ibrahim IM, Afolabi SO, Adebayo JO (2021) Dual targeting of cytokine storm and viral replication in COVID-19 by plant-derived steroidal pregnanes: an in silico perspective. Comput Biol Med 134:104406
Meng XY, Zhang HX, Mezei M, Cui M (2011) Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. ;7:146 – 57.10.2174/157340911795677602
Pinzi L, Rastelli G (2019) Molecular Docking: Shifting Paradigms in Drug Discovery. Int J Mol Sci. ;20.10.3390/ijms20184331
Stanzione F, Giangreco I, Cole JC (2021) Chapter four - use of molecular docking computational tools in drug discovery. In: Witty DR, Cox B (eds) Progress in Medicinal Chemistry, vol 60. Elsevier, pp 273–343
Hines M, Blum J (1979) Bend propagation in flagella. II. Incorporation of dynein cross-bridge kinetics into the equations of motion. Biophys J 25:421–441
Ordentlich A, Barak D, Kronman C, Flashner Y, Leitner M, Segall Y et al (1993) Dissection of the human acetylcholinesterase active center determinants of substrate specificity. Identification of residues constituting the anionic site, the hydrophobic site, and the acyl pocket. J Biol Chem 268:17083–17095
Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L et al (1991) Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science 253:872–879
Ashani Y, Grunwald J, Kronman C, Velan B, Shafferman A (1994) Role of tyrosine 337 in the binding of huperzine A to the active site of human acetylcholinesterase. Mol Pharmacol 45:555–560
Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C et al (1993) Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proceedings of the National Academy of Sciences. ;90:9031-5
Colletier JP, Fournier D, Greenblatt HM, Stojan J, Sussman JL, Zaccai G et al (2006) Structural insights into substrate traffic and inhibition in acetylcholinesterase. EMBO J 25:2746–2756
Darvesh S, Hopkins DA, Geula C (2003) Neurobiology of butyrylcholinesterase. Nature Reviews Neuroscience. ;4:131 – 8.10.1038/nrn1035
Xu Y, Li M, Greenblatt H, Chen W, Paz A, Dym O et al (2012) Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations. Acta Crystallogr Sect D: Biol Crystallogr 68:13–25
Mouchlis VD, Melagraki G, Zacharia LC, Afantitis A (2020) Computer-aided drug design of β-secretase, γ-secretase and anti-tau inhibitors for the discovery of novel Alzheimer’s therapeutics. Int J Mol Sci 21:703
Gupta S, Parihar D, Shah M, Yadav S, Managori H, Bhowmick S et al (2020) Computational screening of promising beta-secretase 1 inhibitors through multi-step molecular docking and molecular dynamics simulations-pharmacoinformatics approach. J Mol Struct 1205:127660
Kumar A, Roy S, Tripathi S, Sharma A (2016) Molecular docking based virtual screening of natural compounds as potential BACE1 inhibitors: 3D QSAR pharmacophore mapping and molecular dynamics analysis. J Biomol Struct Dynamics 34:239–249
Ferreira LG, Dos Santos RN, Oliva G, Andricopulo AD (2015) Molecular docking and structure-based drug design strategies. Molecules 20:13384–13421
Lavecchia A (2015) Machine-learning approaches in drug discovery: methods and applications. Drug Discovery Today 20:318–331
Karplus M, McCammon JA (2002) Molecular dynamics simulations of biomolecules. Nat Struct Biol 9:646–652
Cheng X, Ivanov I (2012) Molecular dynamics. Computational toxicology. :243 – 85
Dong Y-w, Liao M-l, Meng X-l, Somero GN (2018) Structural flexibility and protein adaptation to temperature: Molecular dynamics analysis of malate dehydrogenases of marine molluscs. Proceedings of the National Academy of Sciences. ;115:1274-9
Sinha S, Wang SM (2020) Classification of VUS and unclassified variants in BRCA1 BRCT repeats by molecular dynamics simulation. Comput Struct Biotechnol J 18:723–736
Perez A, Morrone JA, Simmerling C, Dill KA (2016) Advances in free-energy-based simulations of protein folding and ligand binding. Curr Opin Struct Biol 36:25–31
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L et al (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897
Khare G, Andrew Symons R, Do D (2008) Common ophthalmic emergencies. Int J Clin Pract 62:1776–1784
Inestrosa NC, Alvarez A, Perez CA, Moreno RD, Vicente M, Linker C et al (1996) Acetylcholinesterase accelerates assembly of amyloid-β-peptides into Alzheimer’s fibrils: possible role of the peripheral site of the enzyme. Neuron 16:881–891
Guo H, Cao H, Cui X, Zheng W, Wang S, Yu J et al (2019) Silymarin’s inhibition and treatment effects for Alzheimer’s disease. Molecules 24:1748
Haddadi R, Shahidi Z, Eyvari-Brooshghalan S (2020) Silymarin and neurodegenerative diseases: therapeutic potential and basic molecular mechanisms. Phytomedicine 79:153320
Remya C, Dileep K, Tintu I, Variyar E, Sadasivan C (2014) Flavanone glycosides as acetylcholinesterase inhibitors: computational and experimental evidence. Indian J Pharm Sci 76:567
de Almeida P, Boleti APdA, Rüdiger AL, Lourenço GA, da Veiga Junior VF, Lima ES (2015) Anti-inflammatory activity of triterpenes isolated from Protium paniculatum oil-resins. Evidence-Based Complementary and Alternative Medicine. ;2015
Gurovic MSV, Castro MJ, Richmond V, Faraoni MB, Maier MS, Murray AP (2010) Triterpenoids with acetylcholinesterase inhibition from Chuquiraga erinacea D. Don. subsp. erinacea (Asteraceae). Planta Med 76:607–610
Funding
The work received not external funding.
Author information
Authors and Affiliations
Contributions
Conceptualization: Gideon A. Gyebi; Methodology: Gideon A. Gyebi, Oludare M. Ogunyemi, Ibrahim M. Ibrahim; Formal analysis and investigation: Gideon A. Gyebi; Writing - original draft preparation: Gideon (A) Gyebi; Writing - review and editing: Olalekan (B) Ogunro, Saheed O. Afolabi, Gabriel O. Anyanwu, Ibrahim M. Ibrahim, Rotimi J. Ojo; Resources: Gaber El-Saber Batiha; Supervision: Joseph O. Adebayo.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no potential conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gyebi, G.A., Ogunyemi, O.M., Ibrahim, I.M. et al. Identification of potential inhibitors of cholinergic and β-secretase enzymes from phytochemicals derived from Gongronema latifolium Benth leaf: an integrated computational analysis. Mol Divers (2023). https://doi.org/10.1007/s11030-023-10658-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-023-10658-y