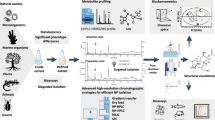
Abstract
Introduction
Molecular networking (MN) has emerged as a key strategy to organize and annotate untargeted tandem mass spectrometry (MS/MS) data generated using either data independent- or dependent acquisition (DIA or DDA). The latter presents a time-efficient approach where full scan (MS1) and MS2 spectra are obtained with shorter cycle times. However, there are limitations related to DDA parameters, some of which are (i) intensity threshold and (ii) collision energy. The former determines ion prioritization for fragmentation, and the latter defines the fragmentation of selected ions. These DDA parameters inevitably determine the coverage and quality of spectral data, which would affect the outputs of MN methods.
Objectives
This study assessed the extent to which the quality of the tandem spectral data relates to MN topology and subsequent implications in the annotation of metabolites and chemical classification relative to the different DDA parameters employed.
Methods
Herein, characterising the metabolome of Momordica cardiospermoides plants, we employ classical MN performance indicators to investigate the effects of collision energies and intensity thresholds on the topology of generated MN and propagated annotations.
Results
We demonstrated that the lowest predefined intensity thresholds and collision energies result in comprehensive molecular networks. Comparatively, higher intensity thresholds and collision energies resulted in fewer MS2 spectra acquisition, subsequently fewer nodes, and a limited exploration of the metabolome through MN.
Conclusion
Contributing to ongoing efforts and conversations on improving DDA strategies, this study proposes a framework in which multiple DDA parameters are utilized to increase the coverage of ions acquired and improve the global coverage of MN, propagated annotations, and the chemical classification performed.





Similar content being viewed by others
Data Availability
Not applicable.
References
Afoullous, S., Balsam, A., Allcock, A. L., & Thomas, O. P. (2022). Optimization of LC-MS2 data acquisition parameteres for molecular networking applied to marine natural products. Metabolites, 12(3), 245. https://doi.org/10.3390/metabo12030245.
Allard, P. M., Péresse, T., Bisson, J., Gindro, K., Marcourt, L., Pham, V. C., et al. (2016). Integration of molecular networking and in-silico ms/ms fragmentation for natural products dereplication. Analytical Chemistry, 88(6), 3317–3323. https://doi.org/10.1021/acs.analchem.5b04804.
Allen, F., Greiner, R., & Wishart, D. (2015). Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics, 11(1), 98–110. https://doi.org/10.1007/s11306-014-0676-4.
Aron, A. T., Gentry, E. C., McPhail, K. L., Nothias, L. F., Nothias-Esposito, M., Bouslimani, A., et al. (2020). Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nature Protocols, 15(6), 1954–1991. https://doi.org/10.1038/s41596-020-0317-5.
Bai, Y., Jia, Q., Su, W., Yan, Z., Situ, W., He, X. (2020). Integration of molecular networking and fingerprint analysis for studying constituents in Microctis Folium. PLoS ONE, 15(7), e0235533. https://doi.org/0.1371/journal.pone.0235533
Barbier Saint Hilaire, P., Rousseau, K., Seyer, A., Dechaumet, S., Damont, A., Junot, C., & Fenaille, F. (2020). Comparative evaluation of data dependent and data independent acquisition workflows implemented on an orbitrap fusion for untargeted metabolomics. Metabolites, 10(4), 158. https://doi.org/10.3390/metabo10040158.
Blaženović, I., Kind, T., Ji, J., & Fiehn, O. (2018). Software tools and approaches for compound identification of lc-ms/ms data in metabolomics. Metabolites, 8(2), 31. https://doi.org/10.3390/metabo8020031.
Bortolotti, M., Mercatelli, D., & Polito, L. (2019). Momordica charantia, a nutraceutical approach for inflammatory related diseases. Frontiers in Pharmacology, 10(MAY), 1–9. https://doi.org/10.3389/fphar.2019.00486.
Chen, Q., Zhang, Y., Zhang, W., & Chen, Z. (2011). Identification and quantification of oleanolic acid and ursolic acid in chinese herbs by liquid chromatography-ion trap mass spectrometry. Biomedical Chromatography, 25(12), 1381–1388. https://doi.org/10.1002/bmc.1614.
Da Silva, R. R., Dorrestein, P. C., & Quinn, R. A. (2015). Illuminating the dark matter in metabolomics. Proceedings of the National Academy of Sciences of the United States of America, 112(41), 12549–12550. https://doi.org/10.1073/pnas.1516878112.
Davies, V., Wandy, J., Weidt, S., Van Der Hooft, J. J. J., Miller, A., Daly, R., & Rogers, S. (2021). Rapid development of improved data-dependent acquisition strategies. Analytical Chemistry, 93(14), 5676–5683. https://doi.org/10.1021/acs.analchem.0c03895.
Defossez, E., Bourquin, J., Reuss, S., Rasmann, S., & Glauser, G. (2021). Eight key rules for successful data-dependent acquisition in mass spectrometry‐based metabolomics. Mass Spectrometry Reviews, (April), mas.21715. https://doi.org/10.1002/mas.21715
Dührkop, K., Nothias, L. F., Fleischauer, M., Reher, R., Ludwig, M., Hoffmann, M. A., et al. (2021). Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nature Biotechnology, 39(4), 462–471. https://doi.org/10.1038/s41587-020-0740-8.
Dührkop, K., Shen, H., Meusel, M., Rousu, J., & Böcker, S. (2015). Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proceedings of the National Academy of Sciences, 112(41), 12580–12585. https://doi.org/10.1073/pnas.1509788112
Ernst, M., Kang, K., Bin, Caraballo-Rodríguez, A. M., Nothias, L. F., Wandy, J., Chen, C., et al. (2019). MolNetEnhancer: enhanced molecular networks by integrating metabolome mining and annotation tools. Metabolites, 9(7), 144. https://doi.org/10.3390/metabo9070144.
Fenaille, F., Saint-Hilaire, B., Rousseau, P., K., & Junot, C. (2017). Data acquisition workflows in liquid chromatography coupled to high resolution mass spectrometry-based metabolomics: where do we stand? Journal of Chromatography A, 1526(March), 1–12. https://doi.org/10.1016/j.chroma.2017.10.043.
Gillet, L. C., Navarro, P., Tate, S., Röst, H., Selevsek, N., Reiter, L., et al. (2012). Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Molecular and Cellular Proteomics, 11(6), 1–17. https://doi.org/10.1074/mcp.O111.016717.
Guo, J., & Huan, T. (2020). Comparison of full-scan, data-dependent, and data-independent acquisition modes in liquid chromatography-mass spectrometry based untargeted metabolomics. Analytical Chemistry, 92(12), 8072–8080. https://doi.org/10.1021/acs.analchem.9b05135.
Heiles, S. (2021). Advanced tandem mass spectrometry in metabolomics and lipidomics—methods and applications. Analytical and Bioanalytical Chemistry, 413(24), 5927–5948. https://doi.org/10.1007/s00216-021-03425-1.
Heinonen, M., Shen, H., Zamboni, N., & Rousu, J. (2012). Metabolite identification and molecular fingerprint prediction through machine learning. Bioinformatics, 28(18), 2333–2341. https://doi.org/10.1093/bioinformatics/bts437.
Kachlicki, P., Piasecka, A., Stobiecki, M., & Marczak, Ł. (2016). Structural characterization of flavonoid glycoconjugates and their derivatives with mass spectrometric techniques. Molecules, 21(11), 1–21. https://doi.org/10.3390/molecules21111494.
Madala, N. E., Tugizimana, F., & Steenkamp, P. A. (2014). Development and optimization of an UPLC-QTOF-MS/MS method based on an in-source collision induced dissociation approach for comprehensive discrimination of chlorogenic acids isomers from Momordica plant species. Journal of Analytical Methods in Chemistry, 2014. https://doi.org/10.1155/2014/650879
Madala, N., Edwin, Piater, L., Dubery, I., & Steenkamp, P. (2016). Distribution patterns of flavonoids from three Momordica species by ultra-high performance liquid chromatography quadrupole time of flight mass spectrometry: a metabolomic profiling approach. Revista Brasileira de Farmacognosia, 26(4), 507–513. https://doi.org/10.1016/j.bjp.2016.03.009.
Makita, C., Chimuka, L., Steenkamp, P., Cukrowska, E., & Madala, E. (2016). Comparative analyses of flavonoid content in Moringa oleifera and Moringa ovalifolia with the aid of UHPLC-qTOF-MS fingerprintin. South African Journal of Botany, 105, 116–122. https://doi.org/10.1016/j.sajb.2015.12.007.
Musharraf, S. G., Kanwal, N., & Arfeen, Q. (2013). Stress degradation studies and stability-indicating TLC-densitometric method of glycyrrhetic acid. Chemistry Central Journal, 7(1), 1–10. https://doi.org/10.1186/1752-153X-7-9.
Nagarani, G., Abirami, A., & Siddhuraju, P. (2014). Food prospects and nutraceutical attributes of Momordica species: a potential tropical bioresources – a review. Food Science and Human Wellness, 3(3–4), 117–126. https://doi.org/10.1016/j.fshw.2014.07.001.
Nicolescu, T. O. (2017). Interpretation of mass spectra. In Mass Spectrometry. InTech. https://doi.org/10.5772/intechopen.68595
Nothias, L. F., Petras, D., Schmid, R., Dührkop, K., Rainer, J., Sarvepalli, A., et al. (2020). Feature-based molecular networking in the GNPS analysis environment. Nature Methods, 17(9), 905–908. https://doi.org/10.1038/s41592-020-0933-6.
Novotny, L., Abdel-Hamid, M. E., Hamza, H., Masterova, I., & Grancai, D. (2003). Development of LC-MS method for determination of ursolic acid: application to the analysis of ursolic acid in Staphylea holocarpa Hemsl. Journal of Pharmaceutical and Biomedical Analysis, 31(5), 961–968. https://doi.org/10.1016/S0731-7085(02)00706-9.
Pilon, A. C., Gu, H., Raftery, D., Bolzani, V. S., Lopes, N. P., Castro-Gamboa, I., & Carnevale Neto, F. (2019). Mass spectral similarity networking and gas-phase fragmentation reactions in the structural analysis of flavonoid glycoconjugates. Analytical Chemistry, 91(16), 10413–10423. https://doi.org/10.1021/acs.analchem.8b05479.
Quinn, R. A., Nothias, L., Vining, O., Meehan, M., Esquenazi, E., & Dorrestein, P. C. (2017). Molecular networking as a drug discovery, drug metabolism, and precision medicine strategy. Trends in Pharmacological Sciences, 38(2), 143–154. https://doi.org/10.1016/j.tips.2016.10.011.
Ramabulana, A., Petras, D., Madala, N. E., & Tugizimana, F. (2021). Metabolomics and molecular networking to characterize the chemical space of four Momordica plant species. Metabolites, 11(11), 763. https://doi.org/10.3390/metabo11110763.
Salem, M. A., De Souza, L. P., Serag, A., Fernie, A. R., Farag, M. A., Ezzat, S. M., & Alseekh, S. (2020, January 15). Metabolomics in the context of plant natural products research: From sample preparation to metabolite analysis. Metabolites. https://doi.org/10.3390/metabo10010037
Scartezzini, P., & Speroni, E. (2000). Review on some plants of indian traditional medicine with antioxidant activity. Journal of Ethnopharmacology, 71(1–2), 23–43. https://doi.org/10.1016/S0378-8741(00)00213-0.
Tebani, A., Afonso, C., & Bekri, S. (2018). Advances in metabolome information retrieval: turning chemistry into biology. Part I: analytical chemistry of the metabolome. Journal of Inherited Metabolic Disease, 41(3), 379–391. https://doi.org/10.1007/s10545-017-0074-y.
Uddin, J., Muhsinah, A., Bin, Imran, M., Khan, M. N., & Musharraf, S. G. (2022). Structure–fragmentation study of pentacyclic triterpenoids using electrospray ionization quadrupole time-of‐flight tandem mass spectrometry (ESI‐QTOFMS/MS). Rapid Communications in Mass Spectrometry, 36(4), https://doi.org/10.1002/rcm.9243.
Vachet, R. W., Winders, A. D., & Glish, G. L. (1996). Correlation of kinetic energy losses in high-energy collision-induced dissociation with observed peptide product ions. Analytical Chemistry, 68(3), 522–526. https://doi.org/10.1021/ac950893r.
van der Laan, T., Boom, I., Maliepaard, J., Dubbelman, A. C., Harms, A. C., & Hankemeier, T. (2020). Data-independent acquisition for the quantification and identification of metabolites in plasma. Metabolites, 10(12), 1–14. https://doi.org/10.3390/metabo10120514.
Vincenti, F., Montesano, C., Di Ottavio, F., Gregori, A., Compagnone, D., Sergi, M., & Dorrestein, P. (2020). Molecular networking: a useful tool for the identification of new psychoactive substances in seizures by LC–HRMS. Frontiers in Chemistry, 8(November), 1–9. https://doi.org/10.3389/fchem.2020.572952.
Wang, M., Carver, J. J., Phelan, V. V., Sanchez, L. M., Garg, N., Peng, Y., et al. (2016). Sharing and community curation of mass spectrometry data with GNPS. Nature Biotechnology, 34(8), 828–837. https://doi.org/10.1038/nbt.3597.Sharing.
Wang, R., Yin, Y., & Zhu, Z. J. (2019). Advancing untargeted metabolomics using data-independent acquisition mass spectrometry technology. Analytical and Bioanalytical Chemistry, 411(19), 4349–4357. https://doi.org/10.1007/s00216-019-01709-1.
Webb, I. K. (2022). Recent technological developments for native mass spectrometry. Biochimica et Biophysica Acta - Proteins and Proteomics, 1870(1), 140732. https://doi.org/10.1016/j.bbapap.2021.140732.
Xu, R., Lee, J., Chen, L., & Zhu, J. (2021). Enhanced detection and annotation of small molecules in metabolomics using molecular-network-oriented parameter optimization. Molecular Omics, 17(5), 665–676. https://doi.org/10.1039/D1MO00005E.
Yan, Z., & Yan, R. (2015). Improved data-dependent acquisition for untargeted metabolomics using gas-phase fractionation with staggered mass range. Analytical Chemistry, 87(5), 2861–2868. https://doi.org/10.1021/ac504325x.
Yang, J. Y., Sanchez, L. M., Rath, C. M., Liu, X., Boudreau, P. D., Bruns, N., et al. (2013). Molecular networking as a dereplication strategy. Journal of Natural Products, 76(9), 1686–1699. https://doi.org/10.1021/np400413s.
Acknowledgements
University of Venda, biochemistry department are gratefully thanked for access to the SHIMADZU LCMS-9030 qTOF. The South African national research fund (NRF) is highly thanked for bursary support to A.-T.R.
Author information
Authors and Affiliations
Contributions
A-TR: Methodology, data analysis, data curation, writing of original draft, writing-review and editing. D.P: supervision, project administration, data curation and analysis. N.E.M: Conceptualization, data analysis, data curation, writing of original draft, writing-review and editing, funding acquisition and project administration. F.T: Conceptualization, methodology, data analysis, data curation, writing of original draft, writing-review and editing, funding acquisition and project administration. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ramabulana, AT., Petras, D., Madala, N.E. et al. Mass spectrometry DDA parameters and global coverage of the metabolome: Spectral molecular networks of momordica cardiospermoides plants. Metabolomics 19, 18 (2023). https://doi.org/10.1007/s11306-023-01981-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-023-01981-4