Abstract
The genus Anthracocystis (Ustilaginales, Ustilaginaceae) was recently reinstated for grass-infecting species of smut fungi that have sori with a peridium composed of mostly fungal cells, filiform or slender columellae, persistent spore balls usually composed of dimorphic spores, and lacking sterile cells between spore balls. In this study, Anthracocystis grodzinskae sp. nov. on Euclasta condylotricha is described and illustrated from the Sudanian savanna biome in Benin (West Africa). The new species is compared with two other smut fungi known on Euclasta condylotricha, namely Sporisorium euclastae and Anthracocystis ischaemoides, in Zambia. It differs from these species in a number of morphological characters that are discussed in detail. The systematic position of A. grodzinskae was investigated in a phylogenetic analysis with a concatenated supermatrix of the internal transcribed spacer (ITS) and large subunit (LSU) regions of ribosomal DNA. The dataset included all representatives of Anthracocystis for which sequences were available in the National Center for Biotechnology Information's (NCBI’s) GenBank and that were linked to reliably identified source specimens, related yeast species, and unnamed yeast strains or environmental sequences. The phylogenetic hypothesis derived from the dataset is intended to serve as a backbone tree for Anthracocystis. 19 ITS and 13 LSU sequences were tracked to represent sequences generated from type specimens (holotypes, isotypes or paratypes). These type sequences are recommended to be deposited in the RefSeq Targeted Loci database. This study provides the first explicit evidence that several asexual species are nested within the Anthracocystis lineage. The yeast sequences were scattered in different subclades of Anthracocystis and none of them could be directly assigned to a teleomorphic species. Only one of these yeast anamorphs was assigned to a species, namely Pseudozyma flocculosa. In line with the current code of nomenclature, and following recent practice of merging yeast species with sexual species under the older generic name, this yeast is recombined into Anthracocystis as A. flocculosa. Additionally, new combinations are proposed for four teliosporic species (Anthracocystis andrewmitchellii, A. christineae, A. kenyana, A. warambiensis).
Similar content being viewed by others
Introduction
The smut genus Anthracocystis Bref. has been described for a causative agent of head smut of millet (Panicum miliaceum L.), Anthracocystis destruens (Schltdl.) Bref. (Brefeld 1912; McTaggart et al. 2012b), but has not been accepted by most smut researchers. Until very recently, it was forgotten or considered synonymous with Sporisorium Ehrenb. ex Link (Vánky 2002). Molecular phylogenetic studies by Stoll et al. (2003, 2005) revealed that Sporisorium species are split into two main lineages. This finding was later confirmed by other molecular analyses (Cunnington et al. 2005; Vánky et al. 2006; Vánky and Lutz 2011; McTaggart et al. 2012a; Shivas et al. 2013; Zhang et al. 2013). These two main lineages included the type species of Sporisorium (S. sorghi Ehrenb. ex Link) and Anthracocystis (A. destruens), respectively. In consequence, McTaggart et al. (2012c) reinstated Anthracocystis for the lineage containing A. destruens, and provided diagnostic characters to separate Sporisorium and Anthracocystis. Thus, the most important diagnostic characters of the resurrected and emended genus Anthracocystis are: a peridium composed of mostly fungal cells, filiform or slender columellae, persistent spore balls usually composed of dimorphic spores (i.e., morphologically different inner and outer spores), and a lack of sterile cells between spore balls.
The members of Anthracocystis are predominantly tropical species, with several species occurring in warm temperate regions, and few species introduced together with their hosts to temperate areas. Two species are pathogenic to cultivated crops, i.e., Anthracocystis destruens causing head smut of millet and Anthracocystis ehrenbergii (J.G. Kühn) McTaggart & R.G. Shivas causing long smut of sorghum. Currently, 126 Anthracocystis species are known (McTaggart et al. 2012c; Denchev and Denchev 2013), of which 60 species were reported from Africa (Vánky et al. 2011). The number of Anthracocystis species in Africa may, however, be significantly larger, as this continent has not been sufficiently explored for smut fungi. There are, potentially, many interesting species within continental or regional scales, as was demonstrated by recent findings of smut fungi from herbarium materials and field studies (Piątek 2006, 2009, 2015; Piątek and Vánky 2007; Piątek et al. 2008, 2012, 2014, 2015).
In recent surveys to West Africa, many grasses were screened for smut infections. Euclasta condylotricha (Hochst. ex Steud.) Stapf was continuously smut-disease-free in most of the surveyed locations. In 2012, however, at two places near the southern border of the Pendjari National Park in northern Benin, moderate infections on inflorescences of Euclasta condylotricha were found that were caused by an ovaricolous smut of the genus Anthracocystis. This study aimed to resolve the specific identity of this smut using light and scanning electron microscopy and DNA sequence analyses, and to provide some ecological information obtained from its natural environment.
The phylogenetic placement of this smut fungus was investigated with the internal transcribed spacer (ITS) and large subunit (LSU) regions of ribosomal DNA (rDNA). Sequences from all representatives of Anthracocystis that were available in the National Center for Biotechnology Information's (NCBI’s) GenBank and that were linked to reliably identified source specimens, related yeast species, and unnamed strains or environmental sequences were included in the analyses. Therefore, a second aim of this study was to provide a phylogenetic framework for Anthracocystis that could serve as a backbone tree for future studies on the genus. Additionally, teleomorphic and anamorphic species are unified under one generic name, fulfilling the requirements of the current International Code of Nomenclature for algae, fungi, and plants (ICN).
Materials and methods
Specimen sampling and morphological examination
The specimens examined are listed in Table 1. The herbarium specimens are deposited in KRAM F. The characteristics of sori, spore balls, spores, and peridial cells were studied using dried herbarium material. The specimens were examined by light microscopy (LM) and scanning electron microscopy (SEM). For LM, small pieces of sori were mounted in 80 % lactic acid, heated to boiling and cooled, and then examined under a Nikon Eclipse 80i light microscope. LM micrographs were taken with a Nikon DS-Fi1 camera. 20 spore balls, 50 spores and 10 peridial cells were measured from each specimen, at a magnification of ×1000, using NIS-Elements BR 3.0 imaging software. Except for the walls of spores and peridial cells, the measurements were adjusted to the nearest 0.5 μm. Spore size range, and the mean and standard deviation were calculated for each investigated specimen (Table 1). The species descriptions include the combined values from all measured specimens. For SEM, spore balls with spores were mounted on carbon tabs and fixed to an aluminium stub with double-sided transparent tape. The stubs were sputter-coated with carbon using a Cressington sputter-coater and viewed under a Hitachi S-4700 scanning electron microscope, with a working distance of ca. 12–13 mm. SEM micrographs were taken in the Laboratory of Field Emission Scanning Electron Microscopy and Microanalysis at the Institute of Geological Sciences of the Jagiellonian University, Kraków (Poland).
DNA extraction, polymerase chain reaction (PCR), and sequencing
Genomic DNA was isolated directly from herbarium specimens. For methods of isolation and crushing of fungal material, DNA extraction, amplification of the ITS 1 and ITS 2 regions of the rDNA including the 5.8S rDNA (ITS, about 740 bp) and the 5′-end of the nuclear large subunit ribosomal DNA (LSU, about 640 bp), purification of PCR products, sequencing, and processing of the raw data see Lutz et al. (2004, 2012). DNA sequences determined for this study were deposited in GenBank (accession numbers are given in Fig. 1, Tables 1 and 2).
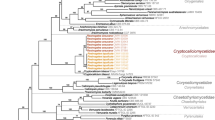
Bayesian inference of phylogenetic relationships within the sampled Anthracocystis species: Markov chain Monte Carlo (MCMC) analysis of an alignment of concatenated ITS + LSU sequences using the GTR + I + G model of DNA substitution with gamma-distributed substitution rates and an estimated proportion of invariant sites, random starting trees and default starting parameters of the DNA substitution model. A 50 % majority-rule consensus tree is shown computed from 75,000 trees that were sampled after the process had become stationary. The topology was rooted to Langdonia confusa and Triodiomyces triodiae. Numbers on branches before slashes are estimates for a posteriori probabilities, numbers on branches after slashes are Maximum Likelihood (ML) bootstrap support values. Branch lengths were averaged over the sampled trees. They are scaled in terms of expected numbers of nucleotide substitutions per site. Sequences in bold are from holotype or isotype specimen (see Table 2). A. = Anthracocystis
Phylogenetic analyses
Blast searches (Altschul et al. 1997) for both the ITS and LSU sequences of the smut specimens on Euclasta condylotricha revealed closest similarity to species of Anthracocystis (sensu McTaggart et al. 2012c) or Pseudozyma Bandoni emend. Boekhout. The phylogenetic position of the smut specimens on Euclasta condylotricha was determined from their concatenated ITS + LSU sequences within a dataset that covered all ITS and LSU sequences of Anthracocystis available in GenBank, as well as all sequences of yeast isolates and uncultured fungus clones that clustered within Anthracocystis. There were two exceptions from the GenBank data that were excluded: doubtful sequences of Sporisorium scitamineum (Syd.) M. Piepenbr., M. Stoll & Oberw. (EF185083, ITS) and Sporisorium nervosum Vánky, C. Vánky & R.G. Shivas (AY740057/AY740110, ITS/LSU). GenBank accession numbers of the sequences used are given in Fig. 1 and Tables 1 and 2.
Sequences were aligned with MAFFT 7.215 applying the L-INS-i option (Katoh and Standley 2013). The alignment was deposited in TreeBASE (S17942). To obtain reproducible results, manipulation of the alignment by hand as well as manual exclusion of ambiguous sites were avoided, as suggested by Giribet and Wheeler (1999) and Gatesy et al. (1993). Instead, highly divergent portions of the alignment were omitted using GBlocks 0.91b (Castresana 2000) with the following options. ‘Minimum Number of Sequences for a Conserved Position’: 33, ‘Minimum Number of Sequences for a Flank Position’: 33, ‘Maximum Number of Contiguous Non-conserved Positions’: 8, ‘Minimum Length of a Block’: 5, and ‘Allowed Gap Positions’ to ‘With half’. The resulting alignment [new number of positions: 1212 (36 % of the original 3362 positions), number of variable sites: 422] was used for phylogenetic analyses using a Maximum Likelihood (ML) and a Bayesian Approach (BA) following Vasighzadeh et al. (2014). In line with the results of McTaggart et al. (2012a), trees were rooted with Langdonia confusa (H.S. Jacks.) McTaggart & R.G. Shivas and Triodiomyces triodiae (Vánky) McTaggart & R.G. Shivas.
Results
Morphological analyses
The two examined specimens on Euclasta condylotricha showed rather inconspicuous, localized smut fungal sori in single ovaries of the inflorescences. The sori were enclosed by whitish peridia, composed mostly of fungal cells, except for a very thin outer layer including the remnants of host epidermis. The sori contained a mass of spore balls surrounding several filiform columellae. The spore balls were composed of dimorphic spores: a single layer of darker and distinctly ornamented outer spores, and numerous lighter and less distinctly ornamented inner spores. The sizes of outer and inner spores and average sizes were uniform and comparable both within each of the collections as well as between the two collections (Table 1). Sterile cells between the spore balls were not observed. The detailed morphological characterization of the smut fungus on Euclasta condylotricha is included in the species description and depicted in Figs. 2, 3 and 4.
Anthracocystis grodzinskae on Euclasta condylotricha: a type locality in Benin, infected plants occurred at the base of the hill, in the transition zone between rocky savanna and gallery forest, b inflorescence of Euclasta condylotricha with five scattered sori (indicated by white arrows) infecting single ovaries, c unopened sorus infecting the ovary, d opened sorus with four visible filiform columellae. Scale bars = 1 cm
Anthracocystis grodzinskae on Euclasta condylotricha seen by light microscopy (a–e from KRAM F-57394, f–h from KRAM F-57395): a–b peridial cells, c–d spore balls, e–f outer spores, median and superficial views, g–h outer and inner spores, median and superficial views. Scale bars: a–b, e–h = 10 μm, c–d = 20 μm
Phylogenetic analyses
The sequences of the smut specimens on Euclasta condylotricha were identical for both the ITS and LSU. The different runs of the BA that were performed and the ML analyses yielded consistent topologies. To illustrate the results, the consensus tree of one run of the BA is presented (Fig. 1). In all analyses, the species determined on the basis of host plants and morphology [Anthracocystis destruens, A. fallax (R.G. Shivas & Cunnington) McTaggart & R.G. Shivas, A. panici-leucophaei (Bref.) McTaggart & R.G. Shivas, A. tumefaciens (McAlpine) McTaggart & R.G. Shivas), or morphology and physiology [Pseudozyma flocculosa (Traquair, L.A. Shaw & Jarvis) Boekhout & Traquair] were inferred with high support values. The smut specimens on Euclasta condylotricha clustered within the sampled Anthracocystis species forming a distinct clade, however, in an unresolved relationship to the remaining Anthracocystis species, the sampled yeast isolates and the uncultured fungus clones.
Taxonomy
New species
Anthracocystis grodzinskae Piątek & M. Lutz, sp. nov. Figs. 2–4
MycoBank # MB 813430
Etymology: Named in honour of Prof. Dr Hab. Krystyna Grodzińska, Polish botanist and ecologist, to mark her eightieth birthday in 2014. Her scientific life and activities were illustrated by Godzik and Zarzycki (2005).
Type: Benin, Atakora Department: near the Tanougou Waterfalls (Chutes de Tanougou), ca. 55 km N of Natitingou, 10°48′22″N, 01°26′16″E, elev. ca. 265 m a.s.l., on Euclasta condylotricha, 29 Oct. 2012, M. Piątek & N.S. Yorou (holotype: KRAM F-57394).
Description: Parasitic on Euclasta condylotricha. Sori in single ovaries of the inflorescence, forming moderately long, cylindrical bodies, 10–15 mm long, 1.5–2 mm wide, at the base partly concealed by floral envelopes (glumes), covered by a white peridium (dirty yellow in dry material) that during maturity ruptures from the apex, revealing a dark brown, semi-agglutinated mass of spore balls surrounding several (up to 4–5) simple, filiform columellae. Peridium composed of an outer thin layer of host epidermis and an inner thin layer of peridial cells of fungal origin. Peridial cells in well-developed, adhering, parallel chains, single cells hyaline, usually long cylindrical to elongated, rarely broadly ellipsoidal, sometimes inflated, (5.0–)6.0–17.0 × (3.5–)4.0–6.0(−7.0) μm, with 1–2 larger and several smaller spherical refractive bodies (lipid drops) in the cytoplasm; wall even, about 0.5 μm thick, smooth. Spore balls dark reddish-brown, under pressure hardly disintegrating into single spores, polyhedraly subglobose, ellipsoidal, irregular or elongated, 50–180 × 50–130 μm, composed of dozens of spores. Spores dimorphic, outer spores yellow brown, globose or subglobose on plane view, usually somewhat polyangular, and collapsed, especially on the free side, flattened on side view and cup-shaped, 10.0–12.0 × (9.0–)9.5–11.5 μm [av. ± SD, 11.1 ± 0.6 × 10.2 ± 0.6 μm, n = 100/2], wall even, ca. 1.0 μm thick, somewhat darker than the rest of the spore, finely punctate in LM, echinulate in SEM, spore profile smooth or finely serrulate; inner spores subhyaline, pale light brown or pale olive-brown, globose or subglobose, usually irregularly polyangular, rugulose, and partly collapsed, 8.5–12.0(−14.0) × (7.5–)8.5–11.0(−12.0) μm [av. ± SD, 10.7 ± 1.0 × 9.7 ± 0.8 μm, n = 100/2], wall even, ca. 1.0 μm thick, very finely punctate in LM (seen with difficulty using DIC), finely verrucose in SEM. Sterile cells absent.
Other specimen examined (paratype):Benin, Atakora Department: near Tanougou village, ca. 27 km NE of Tanguiéta, 10°48′04″N, 01°25′43″E, elev. ca. 265 m a.s.l., on Euclasta condylotricha, 29 Oct. 2012, M. Piątek & N.S. Yorou (KRAM F-57395).
Location, habitat and population size: The two localities of Anthracocystis grodzinskae are located on the northern slopes of the Atakora Mountains in northern Benin, both within the Sudanian savanna biome, containing also Sahelian elements. The direct distance between both localities is 1.15 km. At the type locality, near the Tanougou Waterfalls, infected plants were found in the transition zone between a gallery forest and rocky savanna in a semi-shaded place. The level of infection of plants was small (less than 5 % of inflorescences were infected). At the second locality, near Tanougou village, infected plants were found in grassy Acacia scrubland, in a semi-shaded place. The level of infection of available plant specimens was very small, and only one infected clump was detected. None of the co-occurring different grass species at both localities were infected by Anthracocystis grodzinskae, indicating that this smut is specific to Euclasta condylotricha.
Sporulating time: October (Benin).
New combinations
The results of molecular phylogenetic analyses and/or morphological characters indicate that four Sporisorium species and one Pseudozyma species should be transferred to Anthracocystis. Therefore, we propose the following new combinations:
Anthracocystis andrewmitchellii (R.G. Shivas, McTaggart & Vánky) M. Lutz & Piątek, comb. nov. – MycoBank # MB 813431.
Basionym: Sporisorium andrewmitchellii R.G. Shivas, McTaggart & Vánky, in Crous et al., Persoonia 28: 155 (2012).
Comments: This smut, described on Enneapogon aff. lindleyanus (Domin) C.E. Hubb. from Australia (Crous et al. 2012), is nested inside the Anthracocystis lineage according to the current molecular analyses and its morphology fits the concept of the genus.
Anthracocystis christineae (R.G. Shivas, McTaggart & Vánky) Piątek & M. Lutz, comb. nov. – MycoBank # MB 813437.
Basionym: Sporisorium christineae R.G. Shivas, McTaggart & Vánky, in Vánky et al., Mycol. Balcanica 6: 100 (2009).
Comments: This species was described on Schizachyrium sanguineum (Retz.) Alston from Bolivia (Vánky et al. 2009) and its morphology fits the concept of the genus Anthracocystis.
Anthracocystis flocculosa (Traquair, L.A. Shaw & Jarvis) M. Lutz & Piątek, f.a. comb. nov. – MycoBank # MB 813438
Basionym: Stephanoascus flocculosus Traquair, L.A. Shaw & Jarvis, Can. J. Bot. 66: 927 (1988). – Synonyms: Sporothix flocculosa Traquair, L.A. Shaw & Jarvis, Can. J. Bot. 66: 927 (1988), described as its anamorph; Pseudozyma flocculosa (Traquair, L.A. Shaw & Jarvis) Boekhout & Traquair, J. Gen. Appl. Microbiol. 41: 364 (1995).
Comments: This yeast species, described as foliar epiphyte of Trifolium pratense L. infected by a powdery mildew in Canada (Traquair et al. 1988) and commonly known by its name Pseudozyma flocculosa (Boekhout 1995), is nested inside the Anthracocystis lineage according to the molecular phylogenetic analyses and in line with the present rules of the ICN should be reallocated to this genus.
Anthracocystis kenyana (Piątek) Piątek & M. Lutz, comb. nov. – MycoBank # MB 813439
Basionym: Sporisorium kenyanum Piątek, Polish Bot. J. 51: 160 (2006).
Comments: This species, described on Setaria pallide-fusca (Schumach.) Stapf. & C.E. Hubb. from Kenya (Piątek 2006), is morphologically similar to Anthracocystis panici-leucophaei, and is therefore reallocated to the genus Anthracocystis.
Anthracocystis warambiensis (Piątek & R.G. Shivas) Piątek & M. Lutz, comb. nov. –MycoBank # MB 813440
Basionym: Sporisorium warambiense Piątek & R.G. Shivas, Mycol. Prog. 10: 58 (2011).
Comments: This species was described on Xerochloa laniflora Benth. from Australia (Piątek and Shivas 2011) and its morphology fits the concept of the genus Anthracocystis.
Emendation of Anthracocystis
By inclusion of yeast species, the emendation of the genus Anthracocystis defined previously by characters of teleomorphic species (Brefeld 1912; McTaggart et al. 2012c), is necessary. The generic diagnosis provided by McTaggart et al. (2012c) is included here without alternations and emended by characters placed in bold. The emended part is based on the selected features reported for Anthracocystis flocculosa (syn. Pseudozyma flocculosa; Traquair et al. 1988; Boekhout 1995). The correct assignment of yeast strains to the genus Anthracocystis requires DNA sequence data.
Anthracocystis Bref., Unters. Gesammtgeb. Mykol. 15: 53 (1912), emend. McTaggart & R.G. Shivas, Persoonia 29: 119 (2012), emend. Piątek & M. Lutz, emend. nov.
= Lundquistia Vánky, Mycotaxon 77: 371 (2001). – Type species: Lundquistia fascicularis Vánky.
Teleomorph parasitic on living plants. “Sori replacing inflorescences, all of the racemes or localized in spikelets of an inflorescence. Peridium of vacuolated fungal cells surrounded by a single layer of host cells. Columella composed of vascular bundles surrounded by host parenchyma permeated by inter- and intra-cellular hyphae, often separated into several columellae each around a vascular bundle surrounded by parenchyma, filiform, flexuous, flattened. Sporogenous hyphae coiled. Spores compacted in spore balls, globose to subglobose, often outer spores darker than inner spores. Sterile cells few or absent” (McTaggart et al. 2012c). Anamorph free-living, pseudozyma-like, producing conidiophores and blastoconidia, assimilating myo -inositol, showing positive diazonium blue B and urease reactions, and placed next to the teleomorphic species in the DNA sequence analyses. Type species: Anthracocystis destruens (Schltdl.) Bref.
Discussion
Delineation of Anthracocystis grodzinskae sp. nov.
Macroscopic symptoms and micro-morphological characters observed in the smut specimens attacking Euclasta condylotricha in Benin match well the redefined genus Anthracocystis (McTaggart et al. 2012c). Molecular phylogenetic analyses confirm the morphological assignment, thus stressing the value of the phenotypic characters carved out by McTaggart et al. (2012c) to define and emend the reinstated genus Anthracocystis.
The grass genus Euclasta Franch. includes two tropical, annual species, E. clarkei (Hack.) Cope and E. condylotricha, the latter is distributed in south-western Asia, the former occurs in the grasslands and savannas of neotropical North America and South America, Africa and southern Asia (Clayton et al. 2006). To date, no smut fungi have been reported on E. clarkei, but two different smut fungi were previously found on E. condylotricha, namely Sporisorium euclastae Vánky and Anthracocystis ischaemoides (Henn.) McTaggart & R.G. Shivas. Both of them were recorded on E. condylotricha in Zambia (Vánky 2004) and both are morphologically clearly different from the smut specimens collected on that grass species in Benin, which are accordingly assigned to the novel species Anthracocystis grodzinskae.
Sporisorium euclastae, specific to Euclasta condylotricha and known only from the type locality in Zambia, is a true member of Sporisorium. It differs from the Beninese smut in having sori destroying all raceme-pairs in the inflorescences, one long flagelliform columella, spores of different ornamentation (densely echinulate, clearly visible with SEM comparison), and sterile cells (Vánky 2004). The sterile cells of Sporisorium euclastae would not be confused with the peridial cells of Anthracocystis grodzinskae since they are of different morphology (subglobose, ellipsoidal or irregular, forming irregular groups) and origin (intermixed with spores).
Anthracocystis ischaemoides is parasitic on different species of Hyparrhenia E. Fourn. in Africa (Vánky 2003). However, one smut collection on Euclasta condylotricha from Zambia was also assigned to this species by Vánky (2003, 2004). Anthracocystis ischaemoides differs from A. grodzinskae by having outer spores of different ornamentation (prominently verrucose on the free surface, smooth on the contact sides, visible under SEM) and smooth inner spores (Vánky 2003, 2004). The occurrence of Anthracocystis ischaemoides on Euclasta condylotricha may be wrongly identified and represent a cryptic species with morphological characters that were not observed by Vánky (2003, 2004).
Anthracocystis grodzinskae is described and illustrated, including characters of the peridial cells that are of fungal origin (McTaggart et al. 2012c), and which, with few exceptions, were not often studied in smut fungi. Compared to smuts with descriptions or illustrations of this character, the morphology and manner of peridial cell development in Anthracocystis grodzinskae (cylindrical to elongated cells in adhering, parallelly situated chains) are similar to those illustrated for Anthracocystis cenchri (Lagerh.) McTaggart & R.G. Shivas (Vánky 1985: Fig. 106B, “sterile” cells of Sporisorium cenchri), but different than those described and illustrated for Sporisorium elegantis Vánky (globose, subglobose, broadly ellipsoidal or long cylindrical to elongated cells in more or less well-developed chains or pseudoparenchymatous; Piątek et al. 2014). Thus, the characters of peridial cells may have some diagnostic potential but this should be explored and evaluated in different smut species in future. Another uncommon feature of Anthracocystis grodzinskae are ornamented inner spores. Dimorphic spores are typical for most members of Anthracocystis, but inner spores are usually smooth in most of the species, and are only rarely ornamented, and then with less prominent ornamentation than outer spores (Piątek and Shivas 2011; Vánky 2012).
Phylogenetic framework for Anthracocystis
This study provides the most comprehensive phylogeny of Anthracocystis to date. It is the first phylogeny focused specifically on the genus Anthracocystis, as all previous phylogenetic studies either treated Anthracocystis as a synonym of Sporisorium in a wider context of grass-infecting Ustilaginales (Cunnington et al. 2005; Stoll et al. 2003, 2005; Vánky et al. 2006; Vánky and Lutz 2011; Zhang et al. 2013), or did not show relationships between species (McTaggart et al. 2012a, c; Shivas et al. 2013). The concatenated ITS + LSU dataset was constructed using only sequences that were linked to reliably identified (by smut experts) voucher specimens. ITS and LSU sequences were only concatenated if derived from the same specimen. The doubtful sequences of Sporisorium scitamineum (EF185083) and Sporisorium nervosum (AY740057/AY740110), though nested within the Anthracocystis lineage (data not shown), were not included in the concatenated ITS + LSU dataset. Sporisorium scitamineum EF185083 was probably wrongly identified, as genuine Sporisorium scitamineum is placed in Sporisorium s. str. (Stoll et al. 2005; Vánky et al. 2006; Vánky and Lutz 2011). Sporisorium nervosum AY740057/AY740110 generated from specimen M 56622 (Stoll et al. 2005) may also be wrongly identified, as a sequence of this species (HQ013106), generated from the holotype specimen (BRIP 27019), was nested within the Sporisorium s. str. lineage (McTaggart et al. 2012a). Re-sequencing of both specimens is necessary to resolve which sequences are indeed linked to Sporisorium nervosum.
In the process of assembling the concatenated dataset, it came to light that 18 ITS sequences and 12 LSU sequences of Anthracocystis species, as well as one ITS sequence and one LSU sequence of Triodiomyces triodiae used as the outgroup, were generated from type specimens (holotypes, isotypes or paratypes; Table 2). All but one of these sequences were not annotated in GenBank as type sequences, but through cross-checking the data in GenBank, original publications (see Table 2), respective monographs (Piepenbring 2003; Vánky and Shivas 2008; Vánky 2012), websites (http://www.straininfo.net/strains/336261/browser) or cited herbaria (D. Triebel, pers. comm.) it was possible to link these Anthracocystis sequences to voucher specimens, host plant species, and countries of origin, and, consequently, to track type sequences. The only sequence annotated in GenBank as a type sequence was an ITS sequence of Sporisorium andrewmitchellii in the RefSeq Targeted Loci database. Thus, ITS sequences generated from type specimens and detected in this study are recommended to be deposited in the RefSeq Targeted Loci database (http://www.ncbi.nlm.nih.gov/bioproject/177353; Schoch et al. 2014). Type material information was also provided to the taxonomic curators at NCBI for annotation of corresponding names in the NCBI Taxonomy Browser. This will expand the ability of users to query and BLAST against all GenBank entries tagged with type material by using a phrase, such as “sequence from type[filter]” (for more detailed examples, see Federhen 2014).
In congruence with previous phylogenetic studies (Stoll et al. 2005; Vánky et al. 2006; Vánky and Lutz 2011), the concatenated ITS + LSU dataset resolves species of Anthracocystis, but not the relationships between most of these species. If relationships were resolved, branches were usually weakly supported. For further resolution of relationships between Anthracocystis species, other genes should be analysed, but for routine species identification and generic placement, the current concatenated ITS + LSU dataset suits well. Thus, similar to resolved backbone trees for different phytopathogenic fungi (e.g., Begerow et al. 2002; Hendrichs et al. 2005; Lutz et al. 2005, 2008; Vánky et al. 2008; Maharachchikumbura et al. 2012; Piątek et al. 2013; Hyde et al. 2014; Savchenko et al. 2014; Ariyawansa et al. 2015; McTaggart et al. 2015), the concatenated dataset (alignment deposited in TreeBASE S17942) is intended to serve as a phylogenetic backbone for the genus Anthracocystis for future molecular studies. In addition to currently accepted Anthracocystis species, this study determined that another species, Sporisorium andrewmitchellii (Crous et al. 2012), was nested within Anthracocystis. Furthermore, three other Sporisorium species, S. christineae, S. kenyanum, and S. warambiense (Piątek 2006; Vánky et al. 2009; Piątek and Shivas 2011), for which the type specimens (mainly old material) have not been sequenced, fit the morphological concept proposed for the emended Anthracocystis (McTaggart et al. 2012c). The appropriate nomenclatural combinations for these species are introduced. The genus Anthracocystis currently contains 131 teleomorphic species. Of these, only 38 species (29 %) are represented by ITS (and usually also LSU) sequences in NCBI’s GenBank. Thus, further collecting and sequencing efforts are necessary to provide a complete phylogenetic tree of Anthracocystis.
Yeast sequences nested within Anthracocystis
In addition to teleomorphic species, the Anthracocystis lineage contains several yeast anamorphs well supported within the recovered phylogenetic tree. Sequences were either obtained directly from yeast strains or from environmental samples (as environmental sequences) that likely represent saprobic species. Previously, several phylogenetic studies on yeasts, with limited sampling of teleomorphic species, suggested an affinity of Pseudozyma flocculosa to some species now accommodated in Anthracocystis (e.g., Sampaio 2004; Wang et al. 2006; Seo et al. 2007; Statzell-Tallman et al. 2010; Chamnanpa et al. 2013). However, the current study provides the first explicit evidence that several asexual species are nested within the Anthracocystis lineage using broad sampling of teleomorphic species. Similar to other smut lineages with asexual species (Begerow et al. 2000; Sampaio 2004; Bauer et al. 2005; Boekhout et al. 2006; Nasr et al. 2014), all yeast strains/sequences clustered in different subclades of Anthracocystis and none could be directly assigned to a teleomorphic species. Only one of these yeast anamorphs is assigned to a species, namely Pseudozyma flocculosa (Traquair et al. 1988; Boekhout 1995). In line with the current ICN, this species should be recombined into Anthracocystis, as that generic name is older than Pseudozyma. In addition, the type species of Pseudozyma, P. prolifica Bandoni is phylogenetically distant from Anthracocystis and merely represents the anamorphic stage of Ustilago maydis (DC.) Corda (Sampaio 2004). The ITS and LSU GenBank sequences AB089364/AB089365 used in the concatenated tree (Fig. 1) are generated from the type strain JCM 10321 of Pseudozyma flocculosa (Sugita et al. 2003, confirmed as type strain on the website: http://www.straininfo.net/strains/336261/browser), which justifies the correct placement of this species in Anthracocystis. Therefore, in line with the current code of nomenclature and following recent practice of merging yeast species with sexual species under the older generic name (Selbmann et al. 2014; Yurkov et al. 2015), the recombination of Pseudozyma flocculosa into Anthracocystis is substantiated here. This action has important implications concerning the nomenclature and taxonomy of this model species used as a biocontrol agent against powdery mildews (Avis et al. 2001; Jarvis et al. 2007), and whose genome was recently sequenced (Lefebvre et al. 2013), especially considering that Pseudozyma is a catch-all genus and contains anamorphs linked to different teleomorphic ustilaginalean genera. This is the first step towards merging asexual yeasts with teleomorphic genera in the subphylum Ustilaginomycotina. Anthracocystis flocculosa (syn. Pseudozyma flocculosa) produces an antifungal glycolipid, flocculosin, that is unique for the fungus and responsible for the antibiotic activity against powdery mildews (Mimee et al. 2005; Clément-Mathieu et al. 2008; Teichmann et al. 2011). The evolutionary origin of this fungus, independent of other Pseudozyma species, could, therefore, be mirrored by the production of this distinct glycolipid.
The remaining yeast sequences nested within the Anthracocystis lineage belong to putatively undescribed species, and were generated from strains (or environmental samples) isolated from diverse sources such as soil, rice leaves, sea water, wood, avian guano, bird cloacae or grape berries (Table 2; Mahdi 2006; Waldrop et al. 2006; Francesca et al. 2012; Setati et al. 2012; Takashima et al. 2012). This demonstrates a much wider ecological spectrum of Anthracocystis than revealed by teleomorphic species. Some of these yeast strains were wrongly assigned to known teleomorphic species based on wrongly interpreted similarity in DNA sequences, e.g., the yeast sequences named as “Sporisorium elionuri” (AB863531, AB726473) or “Sporisorium penniseti” (HQ641273). The present phylogenetic analyses revealed these yeast sequences were not related to the teleomorphic species to which they were assigned. This indicates that DNA-based species identification of yeasts in ecological studies by direct comparisons of sequences to the closest hits in GenBank should be done with caution, as it may lead to unreliable interpretations and conclusions. In turn, the development of publicly accessible backbone trees, such as that developed in the current study, will help the phylogenetic placement of yeast anamorphs.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ariyawansa HA, Thambugala KM, Manamgoda DS, Jayawardena R, Camporesi E, Boonmee S, Wanasinghe DN, Phookamsak R, Hongsanan S, Singtripop C, Chukeatirote E, Kang JC, Jones EBG, Hyde KD (2015) Towards a natural classification and backbone tree for Pleosporaceae. Fungal Divers 71:85–139
Avis TJ, Caron SJ, Boekhout T, Hamelin RC, Bélanger RR (2001) Molecular and physiological analysis of the powdery mildew antagonist Pseudozyma flocculosa and related fungi. Phytopathology 91:249–254
Bauer R, Lutz M, Oberwinkler F (2005) Gjaerumia, a new genus in the Georgefischeriales (Ustilaginomycetes). Mycol Res 109:1250–1258
Begerow D, Bauer R, Boekhout T (2000) Phylogenetic placements of ustilaginomycetous anamorphs as deduced from nuclear LSU rDNA sequences. Mycol Res 104:53–60
Begerow D, Lutz M, Oberwinkler F (2002) Implications of molecular characters for the phylogeny of the genus Entyloma. Mycol Res 106:1392–1399
Boekhout T (1995) Pseudozyma Bandoni emend. Boekhout, a genus for yeast-like anamorphs of Ustilaginales. J Gen Appl Microbiol 41:359–366
Boekhout T, Gildemacher P, Theelen B, Müller WH, Heijne B, Lutz M (2006) Extensive colonization of apples by smut anamorphs causes a new postharvest disorder. FEMS Yeast Res 6:63–76
Brefeld O (1912) Die Brandpilze und die Brandkrankheiten. V. Untersuchungen aus dem Gesammtgebiete der Mykologie. XV. Commissions-Verlag von Heinrich Schoningh, Munster
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Chamnanpa T, Limtong P, Srisuk N, Limtong S (2013) Pseudozyma vetiver sp. nov., a novel anamorphic ustilaginomycetous yeast species isolated from the phylloplane in Thailand. Antonie Van Leeuwenhoek 104:637–644
Clayton WD, Vorontsova MS, Harman KT, Williamson H (2006 onwards) GrassBase – The Online World Grass Flora. http://www.kew.org/data/grasses-db.html. Accessed 28 October 2014
Clément-Mathieu G, Chain F, Marchand G, Bélanger RR (2008) Leaf and powdery mildew colonization by glycolipid-producing Pseudozyma species. Fungal Ecol 1:69–77
Crous PW, Summerell BA, Shivas RG, Burgess TI, Decock CA, Dreyer LL, Granke LL, Guest DI, Hardy GESJ, Hausbeck MK, Hüberli D, Jung T, Koukol O, Lennox CL, Liew ECY, Lombard L, McTaggart AR, Pryke JS, Roets F, Saude C, Shuttleworth LA, Stukely MJC, Vánky K, Webster BJ, Windstam ST, Groenewald JZ (2012) Fungal Planet description sheets: 107–127. Persoonia 28:138–182
Cunnington JH, Vánky K, Shivas RG (2005) Lundquistia is a synonym of Sporisorium (Ustilaginomycetes). Mycol Balc 2:95–100
Denchev CM, Denchev TT (2013) Erratomycetaceae, fam. nov., and validation of some names of smut fungi recently described from India. Mycobiota 1:63–70
Federhen S (2014) Type material in the NCBI Taxonomy Database. Nucleic Acids Res 43:D1086–98
Francesca N, Canale DE, Settanni L, Moschetti G (2012) Dissemination of wine-related yeasts by migratory birds. Environ Microbiol Rep 4:105–112
Gatesy J, DeSalle R, Wheeler W (1993) Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Mol Phylogenet Evol 2:152–157
Giribet G, Wheeler WC (1999) On gaps. Mol Phylogenet Evol 13:132–143
Godzik B, Zarzycki K (2005) An appreciation of Professor Krystyna Grodzińska on the occasion of her seventieth birthday. Pol Bot Stud 19:7–26
Hendrichs M, Begerow D, Bauer R, Oberwinkler F (2005) The genus Anthracoidea (Basidiomycota, Ustilaginales): a molecular phylogenetic approach using LSU rDNA sequences. Mycol Res 109:31–40
Hyde KD, Nilsson RH, Alias SA, Ariyawansa HA, Blair JE, Cai L, de Cock AWAM, Dissanayake AJ, Glockling SL, Goonasekara ID, Gorczak M, Hahn M, Jayawardena RS, van Kan JAL, Laurence MH, Lévesque A, Li XH, Liu JK, Maharachchikumbura SSN, Manamgoda DS, Martin FN, McKenzie EHC, McTaggart AR, Mortimer PE, Nair PVR, Pawłowska J, Rintoul TL, Shivas RG, Spies CFJ, Summerell BA, Taylor PWJ, Terhem RB, Udayanga D, Vaghefi N, Walther G, Wilk M, Wrzosek M, Xu JC, Yan JY, Zhou N (2014) One stop shop: backbones trees for important phytopathogenic genera: I (2014). Fungal Divers 67:21–125
Jarvis WR, Traquair JA, Bélanger RR (2007) Perspectives on the development of Sporodex®: fungal biocontrol for powdery mildew in greenhouse crops. In: Goettel VM, Lazarovits G (eds) Biological Control: a Global Perspective. CABI Publishing, UK, pp 224–233
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kellner R, Vollmeister E, Feldbrugge M, Begerow D (2011) Interspecific sex in grass smuts and the genetic diversity of their pheromone-receptor system. PLoS Genet 7(12):E1002436
Lefebvre F, Joly DL, Labbé C, Teichmann B, Linning R, Belzile F, Bakkeren G, Bélanger RR (2013) The transition from a phytopathogenic smut ancestor to an anamorphic biocontrol agent deciphered by comparative whole-genome analysis. The Plant Cell 25:1946–1959
Lutz M, Bauer R, Begerow D, Oberwinkler F, Triebel D (2004) Tuberculina: rust relatives attack rusts. Mycologia 96:614–626
Lutz M, Göker M, Piątek M, Kemler M, Begerow D, Oberwinkler F (2005) Anther smuts of Caryophyllaceae: molecular characters indicate host-dependent species delimitation. Mycol Prog 4:225–238
Lutz M, Piątek M, Kemler M, Chlebicki A, Oberwinkler F (2008) Anther smuts of Caryophyllaceae: molecular analyses reveal further new species. Mycol Res 112:1280–1296
Lutz M, Vánky K, Piątek M (2012) Shivasia gen. nov. for the Australasian smut Ustilago solida that historically shifted through five different genera. IMA Fungus 3:143–154
Maharachchikumbura SSN, Guo LD, Cai L, Chukeatirote E, Wu WP, Sun X, Crous PW, Bhat DJ, McKenzie EHC, Bahkali AH, Hyde KD (2012) A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Divers 56:95–129
Mahdi LE (2006) A survey of marine fungi and yeast. Thesis (M.S.), University of Hawaii at Manoa
McTaggart AR, Doungsa-ard C, Geering ADW, Aime MC, Shivas RG (2015) A co-evolutionary relationship exists between Endoraecium (Pucciniales) and its Acacia hosts in Australia. Persoonia 35:50–62
McTaggart AR, Shivas RG, Geering ADW, Callaghan B, Vánky K, Scharaschkin T (2012a) Soral synapomorphies are significant for the systematics of the Ustilago-Sporisorium-Macalpinomyces complex (Ustilaginaceae). Persoonia 29:63–77
McTaggart AR, Shivas RG, Geering ADW, Vánky K, Scharaschkin T (2012b) A review of the Ustilago-Sporisorium-Macalpinomyces complex. Persoonia 29:55–62
McTaggart AR, Shivas RG, Geering ADW, Vánky K, Scharaschkin T (2012c) Taxonomic revision of Ustilago, Sporisorium and Macalpinomyces. Persoonia 29:116–132
Mimee B, Labbé C, Pelletier R, Bélanger RR (2005) Antifungal activity of flocculosin, a novel glycolipid isolated from Pseudozyma flocculosa. Antimicrob Agents Chemother 49:1597–1599
Nasr S, Soudi MR, Fazeli SAS, Nguyen HDT, Lutz M, Piątek M (2014) Expanding evolutionary diversity in the Ustilaginomycotina: Fereydouniaceae fam. nov. and Fereydounia gen. nov., the first urocystidalean yeast lineage. Mycol Prog 13:1217–1226
Piątek M (2006) Sporisorium kenyanum, a new smut fungus with long twisted sori on Setaria pallide-fusca in Kenya. Pol Bot J 51:159–164
Piątek M (2009) Two smut fungi on Ischaemum: Sporisorium austroafricanum sp. nova and Tolyposporium bogoriense revisited. Ann Bot Fenn 46:425–430
Piątek M (2015) The identity of Entyloma anadelphiae: reclassification and redescription of leaf and stem smut infecting Anadelphia pumila in Guinea. Phytotaxa 192(1):44–49
Piątek M, Lutz M, Chater AO (2013) Cryptic diversity in the Antherospora vaillantii complex on Muscari species. IMA Fungus 4:5–19
Piątek M, Lutz M, Jacobs A, Villablanca F, Wood AR (2015) Epitypification of Tilletia ehrhartae, a smut fungus with potential for nature conservation, biosecurity and biocontrol. Eur J Plant Pathol 143:151–158
Piątek M, Piątek J, Mossebo DC (2012) Recently discovered collections extend the geographical range of the smut fungus Sphacelotheca polygoni-serrulati to Cameroon and Zambia. Pol Bot J 57:285–293
Piątek M, Piątek J, Yorou NS (2014) Emended description and geographical distribution of Sporisorium elegantis (Ustilaginaceae), a species shared between West Africa and India. Phytotaxa 175(3):148–154
Piątek M, Shivas RG (2011) Sporisorium warambiense sp. nov., a fourth smut fungus on Xerochloa in Australia. Mycol Prog 10:57–60
Piątek M, Vánky K (2007) Ustilago aldabrensis, a new species from Seychelles, and two other smut fungi on Dactyloctenium. Mycol Prog 6:213–219
Piątek M, Vánky K, Mossebo DC, Piątek J (2008) Doassansiopsis caldesiae sp. nov. and Doassansiopsis tomasii: two remarkable smut fungi from Cameroon. Mycologia 100:662–672
Piepenbring M (2003) Smut fungi (Ustilaginomycetes p.p. and Microbotryales, Basidiomycota). Flora Neotropica 86((iv):1–291
Piepenbring M, Begerow D, Oberwinkler F (1999) Molecular sequence data assess the value of morphological characteristics for a phylogenetic classification of species of Cintractia. Mycologia 91:485–498
Piepenbring M, Stoll M, Oberwinkler F (2002) The generic position of Ustilago maydis, Ustilago scitaminea, and Ustilago esculenta (Ustilaginales). Mycol Prog 1:71–80
Roux C, Almaraz T, Durrieu G (1998) Phylogeny of fungi responsible for smut of plants based on ITS sequence analysis. CR Acad Sci Ser III 321:603–609
Sampaio JP (2004) Diversity, phylogeny and classification of basidiomycetous yeasts. In: Agerer R, Blanz P, Piepenbring M (eds) Frontiers in Basidiomycote Mycology. IHW-Verlag, Eching, pp 49–80
Savchenko KG, Carris LM, Castlebury LA, Heluta VP, Wasser SP, Nevo E (2014) Stripe smuts of grasses: one lineage or high levels of polyphyly? Persoonia 33:169–181
Schoch CL, Robbertse B, Robert V, Vu D, Cardinali G, Irinyi L, Meyer W, Nilsson RH, Hughes K, Miller AN, Kirk PM, Abarenkov K, Aime MC, Ariyawansa HA, Bidartondo M, Boekhout T, Buyck B, Cai Q, Chen J, Crespo A, Crous PW, Damm U, De Beer ZW, Dentinger BTM, Divakar PK, Dueñas M, Feau N, Fliegerova K, García MA, Ge ZW, Griffith GW, Groenewald JZ, Groenewald M, Grube M, Gryzenhout M, Gueidan C, Guo L, Hambleton S, Hamelin R, Hansen K, Hofstetter V, Hong SB, Houbraken J, Hyde KD, Inderbitzin P, Johnston PR, Karunarathna SC, Kõljalg U, Kovács GM, Kraichak E, Krizsan K, Kurtzman CP, Larsson KH, Leavitt S, Letcher P M, Liimatainen K, Liu JK, Lodge DJ, Luangsaard JJ, Lumbsch HT, Maharachchikumbura SSN, Manamgoda D, Martín MP, Minnis AM, Moncalvo JM, Mulè G, Nakasone KK, Niskanen T, Olariaga I, Papp T, Petkovits T, Pino-Bodas R, Powell MJ, Raja HA, Redecker D, Sarmiento-Ramirez JM, Seifert KA, Shrestha B, Stenroos S, Stielow B, Suh SO, Tanaka K, Tedersoo L, Tellería MT, Udayanga D, Untereiner WA, Uribeondo JD, Subbarao KV, Vágvölgyi C, Visagie C, Voigt K, Walker DM, Weir BS, Weiß M, Wijayawardene NN, Wingfield MJ, Xu JP, Yang ZL, Zhang N, Zhuang WY, Federhen S (2014). Finding needles in haystacks: linking scientific names, reference specimens and molecular data for Fungi. Database 2014:bau061
Selbmann L, Turchetti B, Yurkov A, Cecchini C, Zucconi L, Isola D, Buzzini P, Onofri S (2014) Description of Taphrina antarctica f.a. sp. nov., a new anamorphic ascomycetous yeast species associated with Antarctic endolithic microbial communities and transfer of four Lalaria species in the genus Taphrina. Extremophiles 18:707–721
Seo HS, Um HJ, Min J, Rhee SK, Cho TJ, Kim YH, Lee J (2007) Pseudozyma jejuensis sp. nov., a novel cutinolytic ustilaginomycetous yeast species that is able to degrade plastic waste. FEMS Yeast Res 7:1035–1045
Setati ME, Jacobson D, Andong UC, Bauer F (2012) The vineyard yeast microbiome, a mixed model microbial map. PLoS One 7(12):E52609
Shivas RG, Cunnington JH, Vánky K (2004) Two new species of Ustilaginomycetes on Chrysopogon fallax from Australia. Fungal Divers 16:147–156
Shivas RG, Lutz M, McTaggart AR, Vánky K (2013) Emended description of Anomalomyces (Ustilaginales), including Anomalomyces yakirrae sp. nov. on Yakirra pauciflora (Poaceae) from Australia. Mycobiota 1:17–24
Statzell-Tallman A, Scorzetti G, Fell JW (2010) Candida spencermartinsiae sp. nov., Candida taylorii sp. nov. and Pseudozyma abaconensis sp. nov., novel yeasts from mangrove and coral reef ecosystems. Int J Syst Evol Microbiol 60:1978–1984
Stoll M, Begerow D, Oberwinkler F (2005) Molecular phylogeny of Ustilago, Sporisorium, and related taxa based on combined analyses of rDNA sequences. Mycol Res 109:342–356
Stoll M, Piepenbring M, Begerow D, Oberwinkler F (2003) Molecular phylogeny of Ustilago and Sporisorium species (Basidiomycota, Ustilaginales) based on internal transcribed spacer (ITS) sequences. Can J Bot 81:976–984
Sugita T, Takashima M, Poonwan N, Mekha N, Malaithao K, Thungmuthasawat B, Prasarn S, Luangsook P, Kudo T (2003) The first isolation of ustilaginomycetous anamorphic yeasts, Pseudozyma species, from patients’ blood and a description of two new species: P. parantarctica and P. thailandica. Microbiol Immunol 47:183–190
Takashima M, Sugita T, Van BH, Nakamura M, Endoh R, Ohkuma M (2012) Taxonomic richness of yeasts in Japan within subtropical and cool temperate areas. PLoS One 7(11):E50784
Teichmann B, Labbé C, Lefebvre F, Bölker M, Linne U, Bélanger RR (2011) Identification of a biosynthesis gene cluster for flocculosin a cellobiose lipid produced by the biocontrol agent Pseudozyma flocculosa. Mol Microbiol 79:1483–1495
Traquair JA, Shaw LA, Jarvis WR (1988) New species of Stephanoascus with Sporothrix anamorphs. Can J Bot 66:926–933
Vánky K (1985) Carpathian Ustilaginales. Symb Bot Ups 24(2):1–309
Vánky K (2002) Illustrated Genera of Smut Fungi, 2nd edn. The American Phytopathological Society, St. Paul, Minnesota
Vánky K (2003) The smut fungi (Ustilaginomycetes) of Hyparrhenia (Poaceae). Fungal Divers 12:179–205
Vánky K (2004) Taxonomical studies on Ustilaginomycetes – 24. Mycotaxon 89(1):55–118
Vánky K (2012) Smut fungi of the world. APS Press, St. Paul, Minnesota
Vánky K, Lutz M (2011) Tubisorus, a new genus of smut fungi (Ustilaginomycetes) for Sorosporium pachycarpum. Mycol Balc 8:129–135
Vánky K, Lutz M, Bauer R (2008) About the genus Thecaphora (Glomosporiaceae) and its new synonyms. Mycol Prog 7:31–39
Vánky K, Lutz M, Shivas RG (2006) Anomalomyces panici, new genus and species of Ustilaginomycetes from Australia. Mycol Balc 3:119–126
Vánky K, Shivas RG (2008) Fungi of Australia: The Smut Fungi. Canberra & CSIRO Publishing, Melbourne, ABRS
Vánky K, Shivas RG, McTaggart AR, Vánky C, Arce WA (2009) Additions to the smut fungi (Ustilaginomycetes) of Bolivia. Mycol Balc 6:99–105
Vánky K, Vánky C, Denchev CM (2011) Smut fungi in Africa – a checklist. Mycol Balc 8:1–77
Vasighzadeh A, Zafari D, Selçuk F, Hüseyin E, Kurşat M, Lutz M, Piątek M (2014) Discovery of Thecaphora schwarzmaniana on Rheum ribes in Iran and Turkey: implications for the diversity and phylogeny of leaf smuts on rhubarbs. Mycol Prog 13:881–892
Waldrop MP, Zak DR, Blackwood CB, Curtis CD, Tilman D (2006) Resource availability controls fungal diversity across a plant diversity gradient. Ecol Lett 9:1127–1135
Wang QM, Jia JH, Bai FY (2006) Pseudozyma hubeiensis sp. nov. and Pseudozyma shanxiensis sp. nov., novel ustilaginomycetous anamorphic yeast species from plant leaves. Int J Syst Evol Microbiol 56:289–293
Yurkov AM, Kachalkin AV, Daniel HM, Groenewald M, Libkind D, de Garcia V, Zalar P, Gouliamova DE, Boekhout T, Begerow D (2015) Two yeast species Cystobasidium psychroaquaticum f.a. sp. nov. and Cystobasidium rietchieii f.a. sp. nov. isolated from natural environments, and the transfer of Rhodotorula minuta clade members to the genus Cystobasidium. Antonie Van Leeuwenhoek 107:173–185
Zhang JZ, Guan PG, Tao G, Ojaghian MR, Hyde KD (2013) Ultrastructure and phylogeny of Ustilago coicis. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 14:336–345
Acknowledgments
We thank Dagmar Triebel (Munich, Germany) for checking the type status of some specimens at the Munich herbarium (M), Conrad L. Schoch (Bethesda, USA) for handling the data related to Anthracocystis in GenBank, and Anna Łatkiewicz (Kraków, Poland) for her help with SEM pictures. This work was supported by the National Science Centre (NCN) of Poland (project no. N N303 414037 for the years 2009–2014).
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Dominik Begerow
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Piątek, M., Lutz, M. & Yorou, N.S. A molecular phylogenetic framework for Anthracocystis (Ustilaginales), including five new combinations (inter alia for the asexual Pseudozyma flocculosa), and description of Anthracocystis grodzinskae sp. nov.. Mycol Progress 14, 88 (2015). https://doi.org/10.1007/s11557-015-1114-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11557-015-1114-3