Genetic identification and hybridization in the seagrass genus Halophila (Hydrocharitaceae) in Sri Lankan waters
- Published
- Accepted
- Received
- Academic Editor
- Robert Toonen
- Subject Areas
- Biodiversity, Genetics, Marine Biology, Plant Science, Taxonomy
- Keywords
- DNA barcoding, Hybridization, ITS, Morphological plasticity, rbcL, Species boundary
- Copyright
- © 2020 Liu et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2020. Genetic identification and hybridization in the seagrass genus Halophila (Hydrocharitaceae) in Sri Lankan waters. PeerJ 8:e10027 https://doi.org/10.7717/peerj.10027
Abstract
Seagrasses, as marine angiosperms, play important roles in coastal ecosystems. With increasing anthropogenic impacts, they are facing dramatic declines on a global scale. Halophila is well-known as a complex taxonomic challenge mainly due to high morphological plasticity. By using only a morphological approach, the genus could be over-split or similar species could be erroneously lumped, thus masking its true biodiversity. In the present study, we incorporated genetic identification with morphological examination to reveal the identity of Halophila plants in southern and northwestern Sri Lankan waters. The nuclear ribosomal internal transcribed spacer (ITS) region and chloroplast ribulose-bisphosphate carboxylase gene (rbcL) were used to identify plants collected from the Gulf of Mannar, Puttalam Lagoon, and Matara, Sri Lanka. Based on genetic identification, H. major (Zoll.) Miquel is reported for the first time from Sri Lanka, which might have been misidentified as H. ovalis in previous literature based on morphology alone. We also observed a first hybridization case of Halophila cross between H. ovalis and H. major. Two potential cryptic species were found, herein designated Halophila sp. 1 (allied to H. minor) and Halophila sp. 2 (closely related to H. decipiens). In order to clarify taxonomic ambiguity caused by morphological plasticity and the low resolution of genetic markers, further comparative phylogenomic approaches might be needed to solve species boundary issues in this genus.
Introduction
Seagrasses, a functional group of marine flowering plants found in coastal areas of the world’s oceans, provide essential habitat for many coastal species and support marine food webs, playing critical roles in the balance of coastal ecosystems and human livelihoods (Mtwana Nordlund et al., 2016). It has been shown that seagrass habitat declined worldwide at a rate of 110 km2 per year between 1980 and 2006 (Waycott et al., 2009). Short et al. (2011) suggested 72 seagrass species needed to be listed in the Red List of International Union for the Conservation of Nature (IUCN) based on global population status. Therefore, there is an urgent need to conduct baseline studies (i.e., diversity, abundance, and distribution) for establishing conservation plans in the future. However, identification based on morphological traits in the genus Halophila is considered to be very challenging since few morphological differences or characteristics exist among closely related species (Kuo et al., 2006). Field ecologists without taxonomic knowledge of this genus may either overestimate or underestimate its true biodiversity (Shimada et al., 2012; Tuntiprapas et al., 2015; Kurniawan et al., 2020).
The genus Halophila comprises approximately 20 species within five sections based on morphological differences (Den Hartog & Kuo, 2007; Kuo et al., 2006). Most species in the genus are in section Halophila, which contains species with a pair of petiolate leaves borne on short, erect lateral shoots (Den Hartog & Kuo, 2007; Kuo et al., 2006). All other species are in sections Microhalophila (H. beccarii), Spinulosae (H. spinulosa), Tricostata (H. tricostata), and Americanae (H. engelmannii and H. baillonis). However, molecular genetic studies propose that H. hawaiiana and H. johnsonii should be treated as conspecific with H. ovalis (Short, Moore & Peyton, 2010). The ITS (internal transcribed spacer) sequence is proven to have great resolution for acting as a genetic barcode for the genus Halophila (Kim et al., 2017). Kim et al. (2017) showed that five major clades can be identified in section Halophila, which has relatively simple phyllotaxy compared to other sections. However, five morphologically similar species cannot be distinguished in the Halophila ovalis complex with ITS: H. ovalis, H. minor, H. hawaiiana, H. johnsonii, and H. ovata. On the other hand, that section’s ITS region is capable of identifying H. decipiens, H. major, H. nipponica, H. okinawensis, H. guidichudii, and H. stipulacea. On the other hand, the rbcL (ribulose-bisphosphate carboxylase) gene of the chloroplast is suggested as a potential barcode region for land plants since it can discriminate among species in approximately 85% of congeneric pair-wise comparisons (Newmaster, Fazekas & Ragupathy, 2006). For seagrasses, the combined use of rbcL and matK (maturase K) genes is recommended by the Consortium for the Barcoding of Life (CBOL). However, neither the resolution of rbcL alone or the combination of rbcL and matK can well resolve the phylogenetic relationship of closely related species within the genus Halophila (Nguyen et al., 2015). Since Halophila species have notoriously great morphological plasticity, ITS and rbcL resolution incorporated with detailed morphological examination should provide valuable insight on the biodiversity of the genus in Sri Lanka.
Fourteen species belonging to six genera (60% of Indo-Pacific bioregion seagrasses) have been recorded in Sri Lanka: Enhalus acoroides, H. beccarii, H. decipiens, H. ovalis, H. ovata, H. minor, H. stipulacea, Thalassia hemprichii, Cymodocea rotundata, C. serrulata, Halodule uninervis, H. pinifolia, Ruppia maritima, and Syringodium isoetifolium (Udagedara et al., 2017). Among these, H. ovalis, H. ovata, and H. minor belong to the H. ovalis complex, which is very difficult to differentiate by morphological traits. Udagedara et al. (2017) have also mentioned that the distribution records of seagrasses in the Sri Lankan coast are extremely limited due to three decades of civil conflict ending in 2009 and a concurrent severe decline in seagrasses. A recent study showed that species composition changed and biodiversity decreased from 1991 to 2013 at Puttalam Lagoon in association with human activities (Ranahewa et al., 2018). Therefore, there is an urgent need to understand the distribution and diversity of seagrasses in Sri Lankan waters before local extinctions occur.
In the present study, we incorporate morphological and genetic analyses on plants belonging to the genus Halophila collected from Sri Lankan waters that are difficult to identify by using either approach alone. With this integration, we attempt to reveal potentially overlooked biodiversity and species distribution.
Materials & Methods
Sampling and phylogenetic analyses
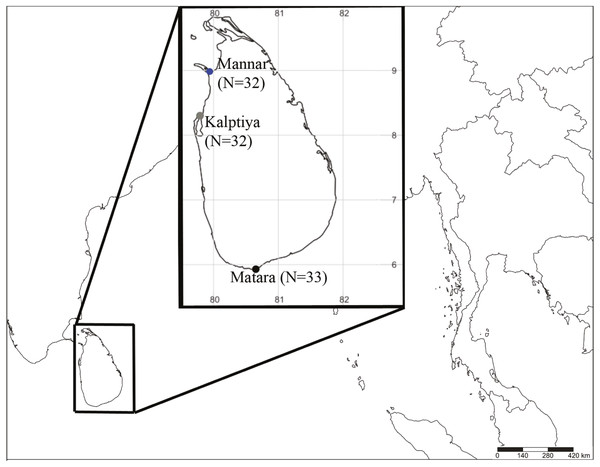
Halophila samples were collected by either snorkeling or sampling from the shoreline in early December 2018 under the permission No. NIC:196931003193 (issued by the Department of Wildlife Conservation) from three sites at depths from 0.5 m to 2 m based on occurrence data in previous literature and information from local fishermen: the Gulf of Mannar (N = 32; GPS: 8.9753739N, 79.9224197E), Puttalam Lagoon, Kalpitiya (N = 32; GPS: 8.3671739N, 79.785565E), and Matara (N = 33; GPS: 5.9474017N, 80.6349298E) (Fig. 1). The former two locations are within a lagoon system having very low visibility (<20 cm). Halophila plants collected from Puttalam were scattered below a huge meadow of Thalassia hemprichii that occurred along the lagoonal side of a sandbar. We collected Halophila plants only from the shoreline at Mannar, since the water is highly polluted by households. Conversely, the site in Matara where we collected Halophila plants faces a coastal ocean area dominated by a 10 x 50 m H. major meadow having >5 m visibility. Leaves were preserved in silica gel for further DNA extraction. Genomic DNA were extracted from leaves using a Plant Genomic DNA Mini Kit (Geneaid Biotech, Taipei, Taiwan). The two markers used in the present study were the rbcL gene from chloroplast DNA and nuclear ITS1-5.8S-ITS2. Primer pairs used in this study were P609 5′-GTAAAATCAAGTCCACCRCG-3′and P610 5′-ATGTCACCACAAACAGAGACTAAAGC-3′ for rbcL (Lucas, Thangaradjou & Papenbrock, 2012) and ITS5a 5′-CCTTATCATTTAGAGGAAGGAG-3′and ITS4 5′-TCCTCCGCTTATTGATATGC-3′ for ITS1-5.8S-ITS2 (Nguyen et al., 2014).
Figure 1: Sample collection sites coded with different colors (blue: Mannar, green: Kalpitiya, and red: Matara) for Halophila surveys in Sri Lanka. Numbers in brackets indicate sample size.
Two loci were amplified in 25 µL reactions in a gradient thermocycler (Veriti 96-well thermal cycler, Thermo Fisher Scientific) over an initial denaturation step at 95 °C for 3 min, 30 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C (ITS1-5.8S-ITS2) and 57 °C (rbcL) for 30 s, extension at 72 °C for 1 min, with a final extension step at 72 °C for 5 min. Each reaction contained 30 ng template DNA, 12.5 ul 2X master Mix RED (15 mM MgCl2 and 0.4 mM each dNTPs), 200 nM of each primer, 0.2 unit of Ampliqon DNA polymerase (Ampliqon, Denmark) and dd water was added to make a final volume of 25 ul. PCR products were sent to Genomics (New Taipei City, Taiwan) for sequencing by an ABI 377 automated sequencer (Carlsbad CA, U.S.A.).
Known ITS sequences and rbcL from other Halophila species were added to the dataset for comparison (Table S1). Sequences obtained in the present study were aligned with reference sequences, including those within sections Spinulosae, Tricostata, Microhalophila, and Americanae as outgroups by MEGA 7 (Kumar, Stecher & Tamura, 2017) to visually inspect all alignments as well as search for the best nucleotide mutation model. Phylogenetic analyses were performed to reveal genetic divergences among Halophila plants collected from different geographic locations, with Bayesian inference assessments through Mr Bayes (MB) version 3.2.2 (Ronquist et al., 2012) and maximum likelihood (ML) being performed by CIPRES Science Gateway (Miller et al., 2015). The former implemented two parallel runs of four simultaneous Markov chains for 10 million generations, sampling every 1000 generations and using default parameters. We discarded the first million generations (10%) as burn-in, based on the stationarity of log-likelihood tree scores. ML analyses were conducted in RAxML version 8.1.24 (Stamatakis, 2014) on CIPRES Science Gateway with default settings. Supporting value on the branches were evaluated by non-parametric bootstrapping with the automatically halt bootstrapping option by RAxML (ML).
Molecular cloning
Among the three sites, a majority of the plants (22/32) collected from Puttalam Lagoon, Kalpitiya, failed to sequence on the ITS1-5.8S-ITS2 region due to multiple templates. We then obtained pure ITS1-5.8S-ITS2 sequences by using molecular cloning.
ITS1-5.8S-ITS2 PCR products amplified from SB21, SB22, SB23, and SB 24 were ligated into pJET1.2/blunt vector and cloned using a CloneJet PCR cloning kit (Thermo Scientific, U.S.A.). In total, 17 positive clones were selected for further PCR reaction. The final cloned PCR fragments were sequenced by Genomics (New Taipei City, Taiwan) using pJET1.2 forward and reverse primers. All sequences derived from the present study were submitted to GenBank under accession numbers MT347850 –MT347937 (ITS) and MT422621 –MT422718 (rbcL).
Morphological analyses
One mature leaf was taken from 48 different plants in each category, comprising H. major, H. sp. 1 (allied to H. minor), H. ovalis, hybrids, potential hybrids, and Halophila sp. 2 (H. decipiens like) (Table 1), for morphological measurements consisting of lamina width, lamina length, distance from intramarginal vein to lamina margin, cross-vein angle, and number of cross-veins (Fig. S1). We calculated the ratio between intramarginal veins to the edge and the half-length of the width, and the ratio between lamina width and length. Specimens were identified using the keys of Den Hartog & Kuo (2007) and Kuo et al. (2006). Morphological data were transformed (x-mean/standard deviation) and subjected to PCA to find out the variation among categories using Past3 software (Hammer, Harper & Ryan, 2001).
| Characteristic | Species | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| H. ovalis | H. major | H. stipulacea | H. decipiens. | Halophila sp2. | |||||||
| Den Hartog (1970) | Halophila sp. 1 (SB) | H. ovalis (SB) | Potential hybrid (SB) | Hybrid (SB) | Kuo et al. (2006) | H. major (MTR) | H. major (MA) | Procaccini et al. (1999)& Vera et al. (2014) | Kuo et al. (2006) | H. decipiens like (MA) | |
| Number of samples | 5 | 2 | 6 | 4 | 13 | 10 | 8 | ||||
| Lamina length (mm) | 10–40 | 4.42–6.61 | 11.00–16.00 | 12.00–20.00 | 23.80–33.00 | 15–25 | 19.80–31.00 | 8.05–31.13 | 63.8–84.3 | 20 | 17.75–40.36 |
| Lamina width (mm) | 5–20 | 1.15–1.89 | 3.90–6.89 | 6.55–9.90 | 8.60–14.13 | 9–11 | 10.50–13.75 | 4.72–14.63 | 6.5–8.43 | 4–6 | 2.75–3.89 |
| No. of pair cross veins | 10–25 | 3–4 | 13 | 11–17 | 18–27 | 14–17 | 11–17 | 12–18 | 11–18 | 6–9 | 8–15 |
| Space between intramarginal veins (mm) | 0.1–0.3 | 0.09–0.17 | 0.29–0.33 | 0.34–0.51 | 0.39–0.54 | 0.2 | 0.50–0.72 | 0.29–0.66 | 0.5 (Ruiz & Ballantine, 2004) | 0.25–3 | 0.24–0.39 |
| Cross-vein angles | 45–60° | 46–68° | 54° | 58–82° | 57–62° | 45–60° | 59–78° | 57–80° | 45–60° | n/a | 57–67° |
| LW:LL | n/a | 0.25–0.35 | 0.43–0.35 | 0.33–0.59 | 0.33–0.43 | n/a | 0.44–0.56 | 0.40–0.59 | n/a | 0.2–0.33 | 0.09–0.20 |
| HLW:DE | 0.5–0.63 | 4.95–10.03 | 10.51–6.75 | 7.90–12.94 | 10.29–13.25 | 0.71–0.33 | 9.01–12.69 | 8.27–6.32 | n/a | 2–3 | 4.89–6.36 |
Notes:
- n/a
-
not available
Results
Phylogenetic analyses
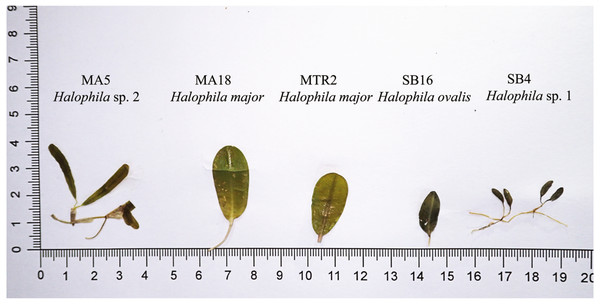
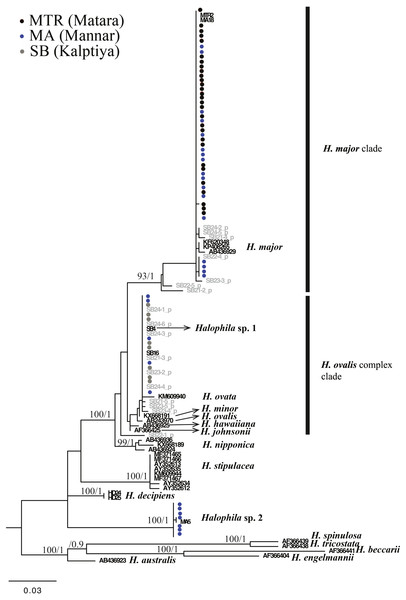
For the ITS1-5.8S-ITS2 region, only 59 of 97 plants were successfully sequenced, 38 failing to achieve consensus sequencing due to the multiple template effect. We subsequently selected four samples for molecular cloning and obtained 17 sequences of the ITS1-5.8S-ITS2 region. In total, with 24 sequences obtained from GenBank and two sequences from H. decipiens collected from southern Taiwan, 14 valid species of Halophila were represented. In total 112 sequences were used for alignment and phylogenetic analyses. The length of aligned sequences is 615 bp, with 157 parsimony informative sites. Based on the ITS phylogenetic tree, sequences in the present study can be divided into three highly supported clades, which are H. major, the H. ovalis complex, and a potential new species (Halophila sp. 2) that is closely related to H. decipiens in terms of anatomic structure as seen under a SEM (J Kuo & S Liu, unpublished data). Interestingly, fresh plants of Halophila sp. 2 are very similar to H. stipulacea (Fig. S2). Outgroups are in sections Microhalophila (H. beccarii), Spinulosae (H. spinulosa), Tricostata (H. tricostata), and Americanae (H. engelmannii), which have complicated phyllotaxy and can clearly be separated from species having simple phyllotaxy in section Halophila, except H. australis. The basal clades of the ingroup comprised three species having leaf edge serrations, including H. stipulacea, H. decipiens, and Halophila sp. (H. stipulacea like clade). Sequences derived from plants collected from all three Sri Lankan sites fell into two main clades: the H. ovalis complex clade and H. major clade. Sequences clustering in the H. major clade were mainly from Mannar and Matara, whereas sequences clustering in the H. ovalis complex clade were from Mannar and Puttalam Lagoon. Interestingly, sequences derived from molecular cloning showed that any given plant contained ITS sequences of both H. ovalis and H. major, as shown in Fig. 2. For example, six sequences were obtained from SB24, two of them belonging to the H. major clade and the remainder to the H. ovalis complex clade. This may indicate a possible hybrid cross between H. ovalis and H. major.
Figure 2: Comparison of leaf morphology of H. major, H. ovalis, and H. stipulacea like specimens collected in Sri Lanka. Samples displayed in this figure were included and showed in ITS tree (Fig. 3).
Figure 3: Phylogeny of Halophila inferred from maximum likelihood and Bayesian analysis based on 615 bp (including gaps) of nrDNA sequences comprising ITS-1, 5.8S rDNA and ITS-2.
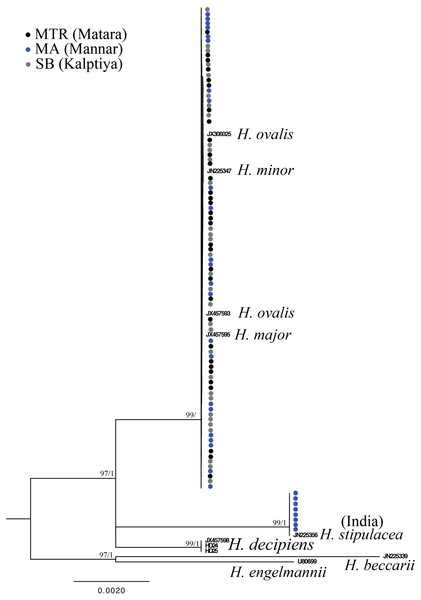
Nodes are presented only for those with bootstrap scores > 90% majority rule for maximum likelihood and > 90% majority probabilities for Bayesian probability values (ML/BI). Sequences are color-coded based on different sampling locations as in Fig. 1. Sample names with shading are sequences derived from molecular cloning.Only one of the 97 samples failed to amplify rbcL. With eight sequences downloaded from GenBank representing seven species of Halophila, the length of the final alignment was 440 bp with only six parsimony informative sites. H. beccarii and H. engelmannii served as outgroups. At the basal-most position of the ingroup, Halophila sp. 2 clustered with a sequence derived from a sample collected from India that was identified as H. stipulacea and sister to H. decipiens. However, sequences derived from most Sri Lankan samples formed a monophyletic clade along with references identified as H. ovalis, H. major, and H. minor (Fig. 3). Since rbcL lacked genetic variation among the different Halophila species, the unresolved phylogeny indicates that rbcL cannot resolve species boundaries in Halophila.
Morphological analyses
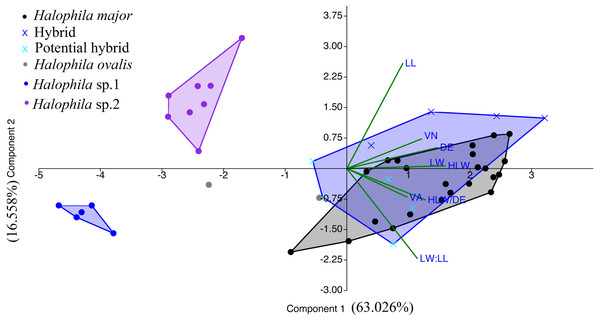
We were unable to measure all the plants that we collected in the present study because we did not collect enough shoots for both DNA extraction and morphological examination during sampling. After DNA extraction, we were able to measure 48 plants, which were further defined into six categories comprised of potential hybrid (failure to sequence without further cloning due to multiple templates), hybrid, H. ovalis, Halophila sp. 1 (allied to H. minor), H. major, and Halophila sp. 2 (H. decipiens like) (Fig. 4). The number of plants examined and results of five measurements and two ratios are given in Table 1. Raw data were transformed and a principal component analysis (PCA) was performed. Variance explained by the first two PCA components (PCA1 and PCA2) is 79.576%. The majority of the variance (95%) of PCA-1 and PCA-2 was explained by lamina length and lamina width:lamina length, respectively. The result of the PCA plot shows that Halophila sp. 1 (allied to H. minor) and Halophila sp. 2 can be distinguished from other categories by smaller lamina length and smaller lamina width:lamina length, respectively. H. major had the widest distribution in the PCA plot, and the hybrid and potential hybrid fell within the range of H. ovalis and H. major (Fig. 5).
Figure 4: Phylogeny of Halophila inferred from maximum likelihood and Bayesian analysis based on 440 bp of the rbcL gene.
Nodes are presented only for those with bootstrap scores > 90% majority rule for maximum likelihood and > 90% majority probabilities for Bayesian probability values (ML/BI). Sequences are color-coded based on different sampling locations as in Fig. 1.Figure 5: Component loadings for the first of two principal components of the PCA of morphological traits with convex hull of different sample groups.
LL, lamina length; LW, lamina width; VA, crossed vein angle; DE, distance between marginal vein and lamina edge; LW/LL, lamina width/lamina length, and HLW/DE, half lamina width/distance between marginal vein and lamina edge.Discussion
The literature related to seagrass communities and biodiversity in the Sri Lanka is scarce. Jayasuriya (1991) mentioned that there are 12 species among nine genera recorded from Sri Lanka, the number being increased to 15 in 2007 (De Silva & Amarasinghe, 2007). The latest report showed 14 species as of 2017 (Udagedara et al., 2017), with the complete list being Enhalus acoroides, Halophila beccarii, H. decipiens, H. ovalis, H. ovata, H. minor, H. stipulacea, Thalassia hemprichii, Cymodocea rotundata, C. serrulata, Halodule uninervis, H. pinifolia, Ruppia maritima, and Syringodium isoetifolium. In the present study, H. major (Zollinger) Miquel (1855) is a new species record for Sri Lanka based on genetic analyses. H. major was previously treated as a synonym of H. ovalis by Den Hartog (1970), and in 2006 Kuo et al. (2006) examined global type materials and concluded that reinstating the taxon status of H. major was warranted. Additionally, further phylogenetic studies (Uchimura et al., 2008; Nguyen, Holzmeyer & Papenbrock, 2013) showed molecular evidence that H. major can also be separated from H. ovalis by using the ITS region. Kuo et al. (2006) suggested that H. major and H. ovalis can be distinguished by a ratio of 1/2 of the lamina width to the distance between the intramarginal veins and lamina margin, and the number of cross-veins. However, most of our measurements of H. ovalis and H. major overlap (Table 1). This morphological plasticity could lead to underestimating seagrass biodiversity as mentioned in Nguyen et al. (2014). In the recent survey of the Gulf of Mannar, Puttalum Lagoon, and southern Sri Lanka (Ranahewa et al., 2018; Ranatunga & Pethiyagoda, 2015; Gunasekara, 2017), H. major may mistakenly be identified as H. ovalis. In addition, our phylogenetic analysis also points out that H. major could be widely distributed in Sri Lanka, since plants collected from Mannar and Matara are identified as H. major. Potential hybrids from Puttalum Lagoon have both H. ovalis and H. major ITS sequences, which may indicate the presence of H. major actually occurring where we failed to collect it. The ITS phylogenetic tree also showed that the H. decipiens like plants (Halophila sp. 2) collected from the Gulf of Mannar are distinct from H. decipiens in Taiwan. Conversely, rbcL sequences derived from H. decipiens like plants cluster with H. stipulacea collected from India. This incongruence could possibly result in erroneous identifications since Halophila sp. 2 resembles H. stipulacea in the field. Another possible explanation is the lack of genetic variation on the rbcL chloroplast gene, which failed to resolve species boundaries in the genus Halophila (Lucas, Thangaradjou & Papenbrock, 2012). The present study also shows that there are only six parsimony informative sites across seven Halophila species among 440 bp.
PCA analyses based on morphology show that Halophila sp.1 (allied to H. minor) and Halophila sp. 2 (H. decipiens like) can be separated from other categories. Although Halophila sp. 2 is very similar to H. stipulacea in appearance, most measurements are smaller than plants in the Mediterranean Sea (Procaccini et al., 1999) as well as those described as H. decipiens in Kuo et al. (2006). As mentioned by Den Hartog (1970), Indian Ocean H. stipulacea plants often have delicate and membranous but never bullate leaves, and more or less deciduous stipules. These plants were initially collected by Isaac Bailey Balfour during the Transit of Venus expedition at Rodrigues Island in 1874 (Balfour, 1879), and later described as Halophila balfourii Soler (Solereder, 1913). Currently, it is treated as a synonym of Halophila stipulacea (Forsskål) Ascherson. Therefore, the plants collected from the Gulf of Mannar could be H. balfourii. However, further genetic analyses from a broad sampling across its current distribution, including the population from the type locality of H. stipulacea and H. decipiens in the Red Sea, is needed to clarify the identity of Halophila sp 2. Most Halophila sp.1 (allied to H. minor) measurements overlap with H. minor or H. ovate (Kuo & Den Hartog, 2001), except that lamina width (1.15–1.89 mm) is smaller compared to these two species (H. minor: 3.5–6 mm; H. ovata: 4-8 mm). Unfortunately, even by using combination loci including matK, rbcL, and trnH-psbA (Lucas, Thangaradjou & Papenbrock, 2012) or ITS, rbcL, and matK (Nguyen et al., 2015), there was a failure to resolve species boundaries in the H. ovalis complex. Further comparative phylogenomic approaches (Liu et al., 2017; Yu et al., 2018) may be useful in resolving H. ovalis complex species boundaries.
Soltis & Soltis (2009) suggested that natural hybridization could be an important creative force and evolutionary process responsible for the increasing of angiosperm species diversity. The incongruence between phylogenetic relationships constructed based on different markers can be considered a signature of hybridization, as well as two divergent alleles of a single locus found in one individual. Intra-species variation in ITS have been identified in many different plant groups, which may hamper attempts to uncover accurate phylogenetic species relationships (Poczai & Hyvönen, 2010). Meanwhile, the high intraspecific variation in ITS is considered as incomplete concerted evolution driven by hybridization (Xu et al., 2017). Additionally, the maternal inheritance of the chloroplast gene tree (i.e., rbcL tree in the present study) reflects only the evolutionary processes of maternal lineages, which may mask genetic evidence of hybridization (Okuyama et al., 2005; Soltis & Soltis, 2009). Either of these reasons may cause incongruence between ITS and plastid phylogenies. Among marine angiosperms, natural hybridization has been observed in only four genera (Halodule, Ruppia, Posidonia, and Zostera) (Ito & Tanaka, 2011; Coyer et al., 2008; Martínez-Garrido et al., 2016; Sinclair, Cambridge & Kendrick, 2019). Ito & Tanaka (2011) found sympatric Halodule uninervis and H. pinifolia hybridizing in the waters of Okinawa by reconstructing their phylogenetic relationship with rbcL and psbA-trnH loci. The congruent pattern between morphological traits and nuclear loci was also observed in two sympatric species of Posidonia (P. australis and P. coriacea) in Australia that show signs of hybridization (Sinclair, Cambridge & Kendrick, 2019). In the present study, the majority of Halophila samples collected from Kapitya failed to sequence due to multiple templates found in single plants, but 17 pure sequences in the ITS region were obtained with further cloning. Phylogenetic analyses showed that a single plant contained ITS sequences clustered with both H. ovalis and H. major (Fig. 2). The percentage of sequencing failure due to multiple templates varied among the three sites (22/32 at Kalpitiya, 1/35 at Mannar, and 5/33 at Matara), indicating that hybridization may be common at these three sites, especially Kalpitiya. However, the PCA plot based on morphology showed that most traits overlap among hybrid, H. ovalis, and H. major. This may be due to the morphological plasticity found in Halophila (Den Hartog & Kuo, 2007; Kuo et al., 2006; Singh, Southgate & Lal, 2019).
Conclusions
In conclusion, Halophila plants collected from Sri Lanka cluster into three clades by the ITS tree, represented as H. major, H. ovalis complex, and Halophila sp. 2 clade. H. major is recorded as a new species of the genus Halophila in Sri Lanka, and may have a wide distribution and possibly be misidentified as H. ovalis in the previous literature. Meanwhile, H. decipiens like plants collected from Mannar may represent a cryptic species of either H. stipulacea or H. decipiens based on phylogenetic relationship traits shared among them. Surprisingly, we found the first case of hybridization in the genus Halophila, which may be a cross between H. ovalis and H. major. Further phylogeographic study with a broader sampling scheme that includes plants from type localities and applying methods based on massive parallel sequencing (i.e., Hyb-Seq, review in Yu et al., 2018) that can obtain genome wide genetic variation is needed to clarify the taxonomic status of Halophila sp. 1 and sp. 2.
Supplemental Information
Illustration of the morphological measurements of the genus Halophila used in present study
List of the reference sequences of Halophila included in the molecular analysis done in this study
Aligned ITS sequences in Fasta format including out-groups
This Fasta file can be viewed by MEGA software.
Aligned rbcL sequences in Fasta format including out-groups
This Fasta file can be viewed by using MEGA software.