Bacterial community characteristics in the rhizosphere of Suaeda glauca versus bulk soil in coastal silt soil modified by sea-sand and their implications
- 1College of Marine Science and Engineering, College of Life Sciences, Nanjing Normal University, Nanjing, China
- 2Lianyungang Golden Coast Development and Construction Co. LTD, Lianyungang, China
In the exploitation and development of the muddy coastal zone in Lianyun New Town, Jiangsu Province, much coastal silt soil (CSS) has been formed through reclamation projects, which is difficult to use. The proportion of wild Suaeda glauca increased significantly and became a dominant species in CSS modified by sea-sand mulching (Treated CSS, TCSS), but it was still affected by saline-alkali stress. To use S. glauca efficiently to improve TCSS, high-throughput sequencing technology and conventional soil analysis methods were adopted to investigate the rhizosphere microbial community structure of S. glauca and the physico-chemical properties of TCSS as well as its internal correlation. The results showed that TCSS had low organic matter content, poor nutrients such as N and P, and poor fertility retention. TCSS was still affected by mild or moderate saline-alkali stress, and the total Ca was greater than 1%, so the soil properties indicated a calcareous saline-alkali soil. The growth of S. glauca can reduce soil salinity and increase soil TN content, and its rhizosphere bacterial community was significantly different from that of bulk soil without S. glauca: the rhizosphere was significantly enriched with C-cycle-related bacterium such as organic matter degrading bacteria mariniflexile, photobacterium Sphingomonadaceae, and N-cycle-related bacterium such as nitrogen-fixing bacteria Azoarcus, denitrifying bacteria Pseudomonadaceae; the symbiotic relationship of rhizosphere bacterial community in S. glauca tended to be more complicated, and the functions of nitrogen-respiration, photo-nutrition, and methyl-nutrition were enhanced. Based on the above results, it is reasonable to predict that the interaction between S. glauca and its rhizosphere bacteria in TCSS could promote the process of soil carbon and nitrogen cycles, as to accelerate soil substance transformation and increase soil fertility, thus driving the benign development of TCSS. The results lay a theoretical basis for developing muddy coasts and promoting the precise improvement of TCSS by large-scale cultivation of S. glauca in the local area.
1. Introduction
The coastal intertidal/supratidal zone is in the ecotone between terrestrial and marine ecosystems (Gómez et al., 2016). As the most sensitive area for energy and material exchange in the atmosphere, water, soil, and biosphere (Von Glasow et al., 2013; Jiang et al., 2015; Ariel et al., 2021), it is one of the most vulnerable areas in terms of being affected by intensive human activities and changes in land use (Murray et al., 2019; Huang et al., 2020a). With the rapid economic development of China’s coastal areas, there is a large influx of people to the eastern coastal zone, and the population growth rate of the eastern coastal provinces reached 10.4% in the last decade (2010-2020), ranking first in China (China 2020 census: Provincial level data and context, https://www.newgeography.com). With the rapid growth of population, the gap in land resources is constantly increasing, which makes the task of improving the saline-alkali soil in the coastal zone extremely urgent. In Jiangsu Province, 97% of the coast is muddy. The soils of these muddy coasts (coastal silt soil, CSS) are classified as intertidal/supratidal silt soils, with deep primary salinization, leading to significant difficulties in the improvement thereof (Zhou, 2015; An et al., 2022). To control the construction cost of soil improvement, a reclamation project in Lianyun New Town, Lianyungang City relied on covering CSS with sea-sand forming a new treated CSS (TCSS) for soil improvement. Although the salt stress level of TCSS decreased significantly, plants still grew sparsely thereafter and were still difficult to use effectively.
The method of soil remediation through plant-microbe interactions is the most popular and efficient green biotechnology. Plant growth can influence soil microbial community composition and diversity by adjusting the soil microenvironment (Zhang et al., 2010; Yan et al., 2018), changing the biological metabolic function of the rhizosphere microenvironment (Chi et al., 2021), further affecting the material cycle and ecological function of the saline-alkali soil environment (Li et al., 2020). Among them, bacteria are the most abundant microbiota in the soil, and they are very diverse in metabolism. Therefore, bacterial diversity is an important biological indicator reflecting environmental disturbance and changes in ecosystem quality (Bouchez et al., 2016; An et al., 2022). Liu et al. (2020) found that the bacterial community related to energy metabolism in the rhizosphere soil of S. salsa growing in intertidal habitats was higher than that in non-rhizosphere soil, while the richness of bacterial community related to carbohydrate and amino acid metabolism decreased; He et al. (2022) restored the biodiversity of the degraded Beidaihe coastal wetland by planting S. salsa, among which the rhizosphere of S. salsa was mainly enriched with dominant bacterial community with functions of aromatic compound degradation; Yuan et al. (2016) found that the genomes of rhizosphere bacteria and endophytic bacteria related to S. salsa were riched in genes that were conducived to adaptation to salt stress, absorption of nutrients, and increased root colonization competitiveness. These studies show that there is functional bacterial community in the rhizosphere of salt-tolerant plants that can help regulate soil material cycling, plant growth, and resist environmental stress. The growth process of salt-tolerant plants not only changes the microbial community structure of the rhizosphere, but also can reduce soil salt-based ion content, improve soil physico-chemical characteristics, and increase soil productivity. For example: Yang et al. (2019) found that S. salsa grows in the soil with total salt content of 233.97 t·ha-1, and its biomass reaches 1035.88 g·m-2 at harvest, which can take away 2.70 t·ha-1 of total salt (including Na+, K+, Ca2+, Mg2+, Cl-, SO42-, NO3-); Lin et al. (2005) found that planting S. salsa can increase soil organic matter by 43% and soil total nitrogen by 18% in saline-alkali soil, while increasing the number of active bacteria and fungi by five and sixteen times, respectively.
The authors have explored wild plants species growing in TCSS in Lianyun New Town since 2018-2021, and found that S. glauca is the only dominant plant specie that can form a single vegetation community in the TCSS, while other plants grow scattered and do not form a distinct community. Therefore, S. glauca was regarded that may be used as the preferred local species for improving TCSS. At present, the dominant microbial community structure of the rhizosphere enrichment of wild S. glauca grown in TCSS and its mechanism of adaptation to the saline-alkali environment remains unclear. We hypothesized that a large number of beneficial bacteria with the function of carbon and nitrogen biotransformation would be enriched by the cultivation of S. glauca, and their interaction with the plant could promote soil improvement. Conducting research in this regard is conducive to the efficient use of the interaction between the rhizosphere bacteria and S. glauca to improve TCSS more effectively.
In the present research, the rhizosphere bacterial diversity and soil physico-chemical properties of wild S. glauca in TCSS were evaluated, the mechanism of S. glauca as a dominant local species in TCSS was explored, and the improvement effect and ecological function of S. glauca growth on TCSS were revealed.
2. Materials and methods
2.1. Description of sampling area and sample collection
The sampling area (within the range of 50m in diameter from the coordinate point E119°27′, N34°79′) lies in the coastal area of Lianyun New Town, Lianyungang City, Jiangsu Province (Figures 1A, B). The soil type is TCSS, which is formed by sea-sand modification project (2015) covering silty sea-sand (to a height of about 0.5 m) on the original coastal silt soil, with a total area of 383.7 ha. Figure 1C demonstrates that S. glauca forms a dominant single vegetative community in this plot. From August to September 2020, bulk soil (TCK) and root-soil of S. glauca (TSG) naturally grown in TCSS were selected as test objects (Figure 1C), with a total of two groups of samples, with three replicates in each group.
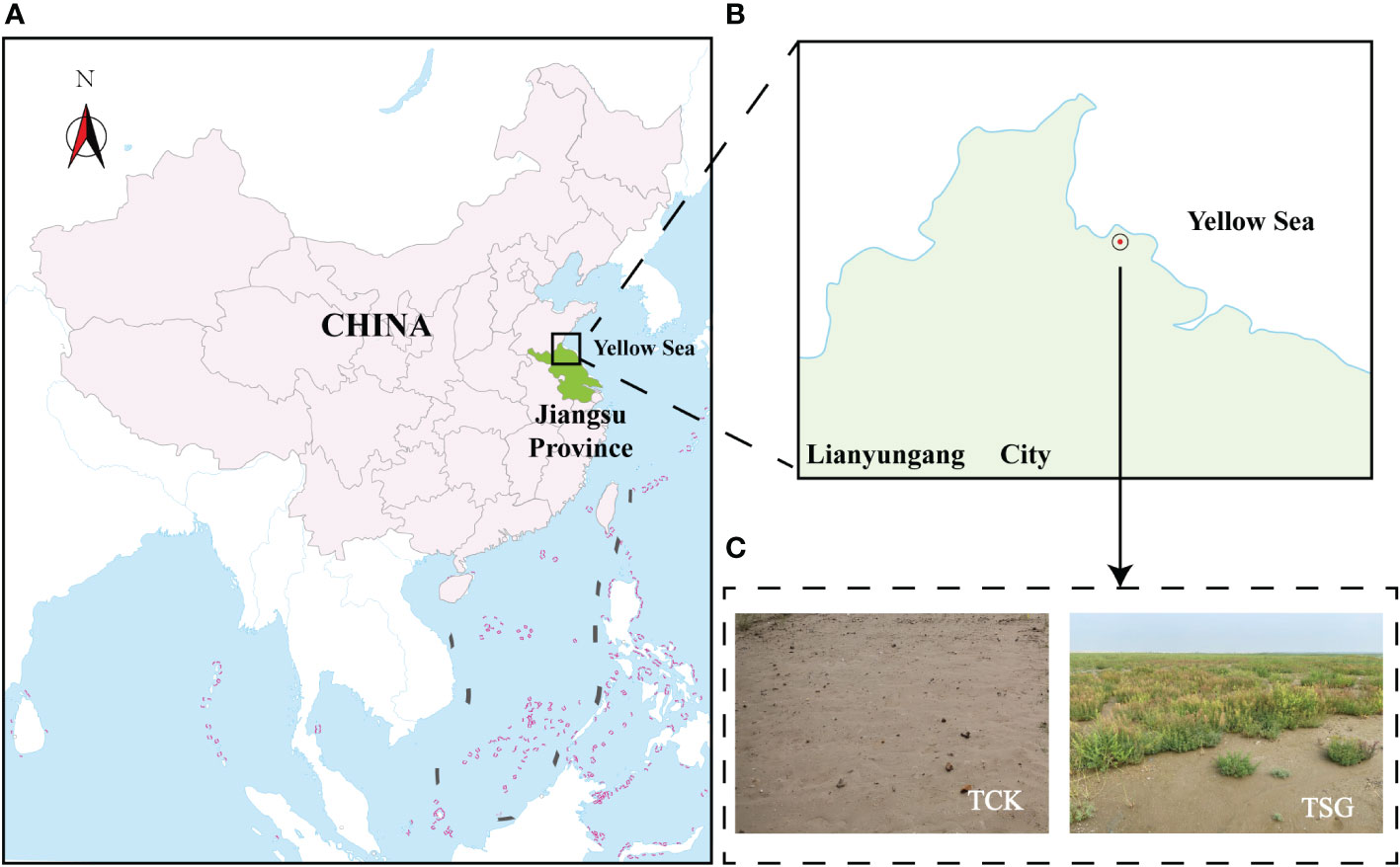
Figure 1 (A, B) Location information of sample plot ((A) It shows the general location information of the sample plot in China; (B) It shows the specific location information of the sampletes plot in Lianyungang City, Jiangsu Province); (C) Photograph showing a TCSS sample plot.
Evenly pull out S. glauca, shake off the bulk soil attached to the roots, and then a sterilized brush was used to place the soil within the root surface of S. glauca into a sterile sampling bag, the collected soil was S. glauca rhizosphere soil (TSG, for DNA extraction); the soil around the S. glauca roots within 10 cm of the root radius (TSG, for determination of soil physico-chemical properties) and the bulk soil without plants growth (TCK, for DNA extraction and determination of physico-chemical properties) was taken with a sterilized shovel, and placed in different sterile sampling bag. The depth of all the collected soil samples ranged between 50 and 150 mm. At the same time, for each repetition of the sample, the soil with the same weight as the five adjacent sampling points was selected and mixed evenly. The collected samples were stored and transported in dry ice, and frozen at -80 °C. In addition, 10 whole plants of S. glauca were excavated for salt-content assay.
In order to show the difference between the CSS (E119°22′, N34°76′) and TCSS, the CSS bulk soil (CK) and the soil around the roots of S. glauca growing on CSS (SG) were collected at the same time, and the collection method was the same as above. The soil samples of CK and SG were collected for the determination of physico-chemical properties, and some of the physico-chemical indexes were published in An et al., 2022.
2.2. Analysis of soil physico-chemical properties and plant salinity
The detection methods of soil physico-chemical properties included: total salt was measured by weighing deionized soil suspension with a soil to water ratio of 1:5 (w/v) (Lv & Li, 2010); electrical conductivity (EC) was measured by conductivity meter (SX731, Shanghai Sanxin, China) using deionized soil suspension with a soil water ratio of 1:2 (w/v); pH measurement by pH meter (PHSJ-4F, Shanghai Leici, China) using deionized soil suspension with a soil to water ratio of 1:2.5 (w/v); cation exchange capacity (CEC) was measured according to Chinese National Standard HJ 889-2017; total nitrogen (TN) was measured according to Chinese National Standard LY/T 1228-2015; total organic carbon (TOC) was determined at 1350 °C by infrared induction instrument (CS844, Leco, US) after pickling soil samples; available potassium (Avail. K) in soil was extracted by neutral ammonium acetate solution (1 mol·L-1), and its content was determined by atomic absorption spectroscopy (AA-6300C, Sgimadzu, Japan); available phosphorus (Avail. P) in soil was extracted by sodium bicarbonate solution, and its content was determined by molybdenum antimony colorimetry (Olsen et al., 1954); total calcium (Total Ca) content of the digestion solution, as soil digested by perchloric acid hydrofluoric acid, was determined by atomic absorption spectroscopy (AA-6300C, Sgimadzu, Japan); available magnesium (Avail. Mg), available calcium (Avail. Ca) and available sodium (Avail. Na) were determined by AB-DTPA extraction solution (Soltanpour, 1985; Malathi & Stalin, 2018) using atomic absorption spectrometry (AA-6300C, Sgimadzu, Japan); the content of water-soluble was determined by EDTA titration (Belle-Oudry, 2008); moisture content (MC) was calculated by “MC (%) = ((weight of wet soil - weight of dry soil)/weight of wet soil) * 100” formula; soil bulk density (SBD) was calculated by “SBD (g·cm-3) = G·100/(V·(100 + MC))” formula, formula “V” denoted the volume of the ring cutter (100 cm3), “G” denoted the weight of wet soil in the ring cutter; specific gravity was determined by using pycnometer (Siebel & Kott, 1937); porosity was calculated by the formula “soil porosity (%) = (1 - soil bulk density/specific gravity) * 100”.
The collected plants were washed and dried at 40 °C, and ground separately according to the above-ground/underground parts (through a 100-mesh sieve), then different parts of the plants were heated and acidized respectively, and the total sodium and total potassium therein were determined using an atomic absorption spectrometer; the content of Cl- was measured according to the Mohr method (Shukla & Arya, 2018).
2.3. Soil total DNA extraction and high-throughput sequencing
Soil total DNA was extracted using the soil DNA extraction kit (MOBIO DNeasy PowerSoil Kit), and the details of the method of extraction were referred to the manufacturer’s instruction. Nossa et al. (2010) showed that 16S rRNA gene V3-V4 hypervariable region bacterial genomic DNA was amplified using primers, V3-V4 universal primers: 343F-5’-TACGGRAGGCAGCAG-3’; 798R-5’-AGGGTATCTAATCCT-3’. High-throughput sequencing was conducted on the Illumina Miseq platform at Shanghai OE Biotech Co., Ltd, China.
2.4. Data processing and statistical analysis
The physico-chemical properties of soil samples were statistically analyzed in SPSS 22.0 (IBM, USA) software and further descriptive, variance, and Pearson correlation analysis were performed. Column graphs of plant salt content were plotted using Origin 8.0 (OriginLab, USA) software.
In order to realize the readability of sequencing data, the following processing will be performed: firstly, the original sequence cuts off the Barcode sequence and Primer sequence by Trimmomatic 0.35 software (Bolger et al., 2014), and then uses Flash 1.2.11 software (Magoč & Salzberg, 2011) to splice the reads of each sample, and the spliced sequence is the original Tags data (Raw Tags); then, Raw Tags uses QIIME 1.8.0 software (Caporaso et al., 2010) for strict filtering to obtain high-quality Tags data (Clean Tags); finally, the UCHIME 2.4.2 software (Edgar et al., 2011) is used to remove the chimeras in the sequence. Vsearch 2.4.2 software was used to cluster the above high-quality screening sequences into operational taxonomic units (OTUs) at a similarity level of 97% (Rognes et al., 2016). The QIIME software was used to select the representative sequences of each OTU, and all the representative sequences would be compared and annotated using the database. The 16S sequence was aligned with the Greengenes database, while species alignment annotation was performed using the RDP classifier software (Wang et al., 2007), and the annotation results with confidence interval greater than 0.7 were obtained.
The indices of bacterial alpha diversity were calculated by using QIIME. Data visualization analysis was carried out using R 3.2.3 software, including drawing of the Venn diagram and community composition column stacking diagram; non-metric multidimensional scaling (NMDS) analysis, anosim tests, and redundancy analysis (RDA) were also performed, as to analyze bacterial community beta diversity and its association with environmental factors; combined use of the WGCNA data package in R and Gephi 0.9.2 software facilitated bacterial co-occurrence network analysis. The bacterial function was predicted using FAPROTAX software.
3. Results
3.1. Analysis of soil physico-chemical properties
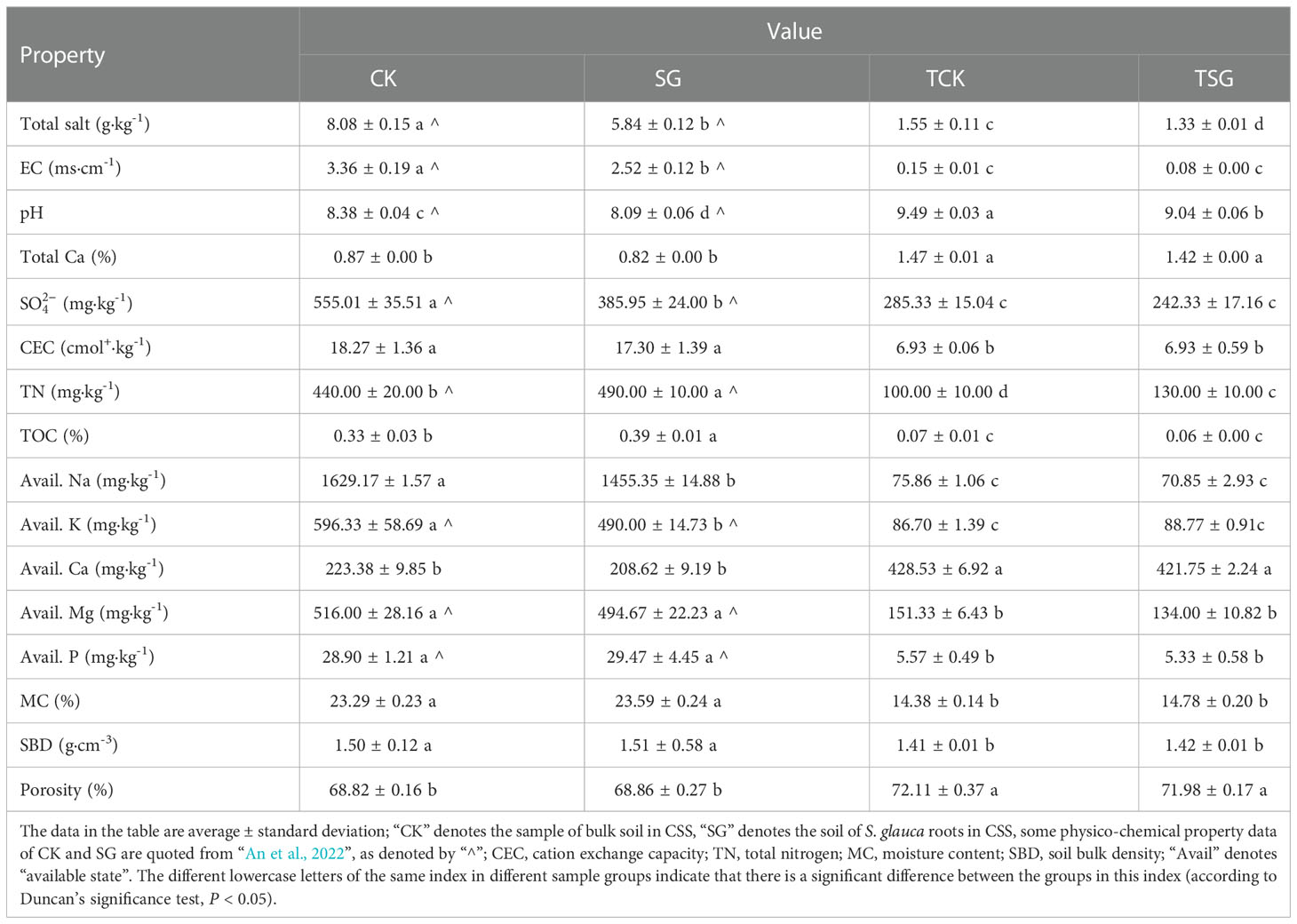
The results of soil physico-chemical index determination are listed in Table 1. Different from the heavily coastal saline-alkali soil properties of bulk soil in CSS (An et al., 2022), the total salt content of bulk soil in TCSS (TCK) is between 1 and 2 g·kg-1, EC and salt-based ion (SO42-, Avail. Na, Avail. K, and Avail. Mg) contents are low; the pH of TCK (≈ 9.5) is relatively high; the fertility indices of CEC, TN, TOC, and Avail. P are poor; MC and SBD are lower, while the porosity is higher; the total Ca content is more than 1%. Pearson correlation analysis (Table S1) of soil physico-chemical properties shows that there is a positive correlation between soil salt-stress-related environmental factors (total salt, EC, SO42-), fertility-related indicators (CEC, TN, TOC), available cations (Avail. Na, Avail. K, Avail. Mg), as well as physical parameters (MC, SBD); the aforementioned environmental factors are negatively correlated with porosity, pH, avail. Ca, and total Ca; porosity, pH, Avail. Ca, and total Ca were positively correlated. The results showed that the alkalinity in the TCSS soil was significantly positively correlated with the soil Ca content; on the other hand, the loss of most salt-based ions (e.g. Na, K, and Mg) and nutrients, is significantly correlated with soil porosity.
Compared with the bulk soil, the trends in the physico-chemical properties of TSG in TCSS were akin to that of SG in CSS (Table 1). The similar trend is such that the physico-chemical properties of the soil around S. glauca roots in both soils showed significant decreasing changes in total salt and pH, and increasing TN content. Among them, the decrease of soil salinity is related to salt absorption by S. glauca. The total amount of salt-based ions (Na+, K+, Cl−) detected in the aboveground and underground parts of S. glauca grown in TCSS, accounted for 16.587% and 8.816% of the dry weight of their respective parts, respectively (Figure S1). The results show that S. glauca grown on TCSS can help soil desalination and increase soil nitrogen content.
3.2. Bacterial diversity analysis
3.2.1. Bacterial diversity and correlation analysis with environmental factors
Alpha diversity represents the diversity of samples, which can represent the diversity of each sample within the habitat, and the results are shown in Figure 2A. According to Chao1, Observed_species, Shannon, and Simpson data, there were no significant differences in bacterial richness and evenness among TCSS samples (TCK, TSG). However, the bacterial diversity of each grouping consisted of different bacterial community structures. The Venn diagram (Figure 2B) showed that the shared OTUs between the two groups only account for 42% of the total OTUs; Beta diversity is displayed by non-metric multidimensional scaling (NMDS) analysis, which reflects the diversity between samples, as shown in Figure 2C. The dimension divides the two groups of samples into two relatively independent regions and being in different regions suggests that there is a certain difference in community composition between samples. Both the Venn diagram and the NMDS analysis results proved that the bacterial community composition in the rhizosphere of S. glauca was significantly different from that in the bulk soil (ANOSIM, P< 0.01).
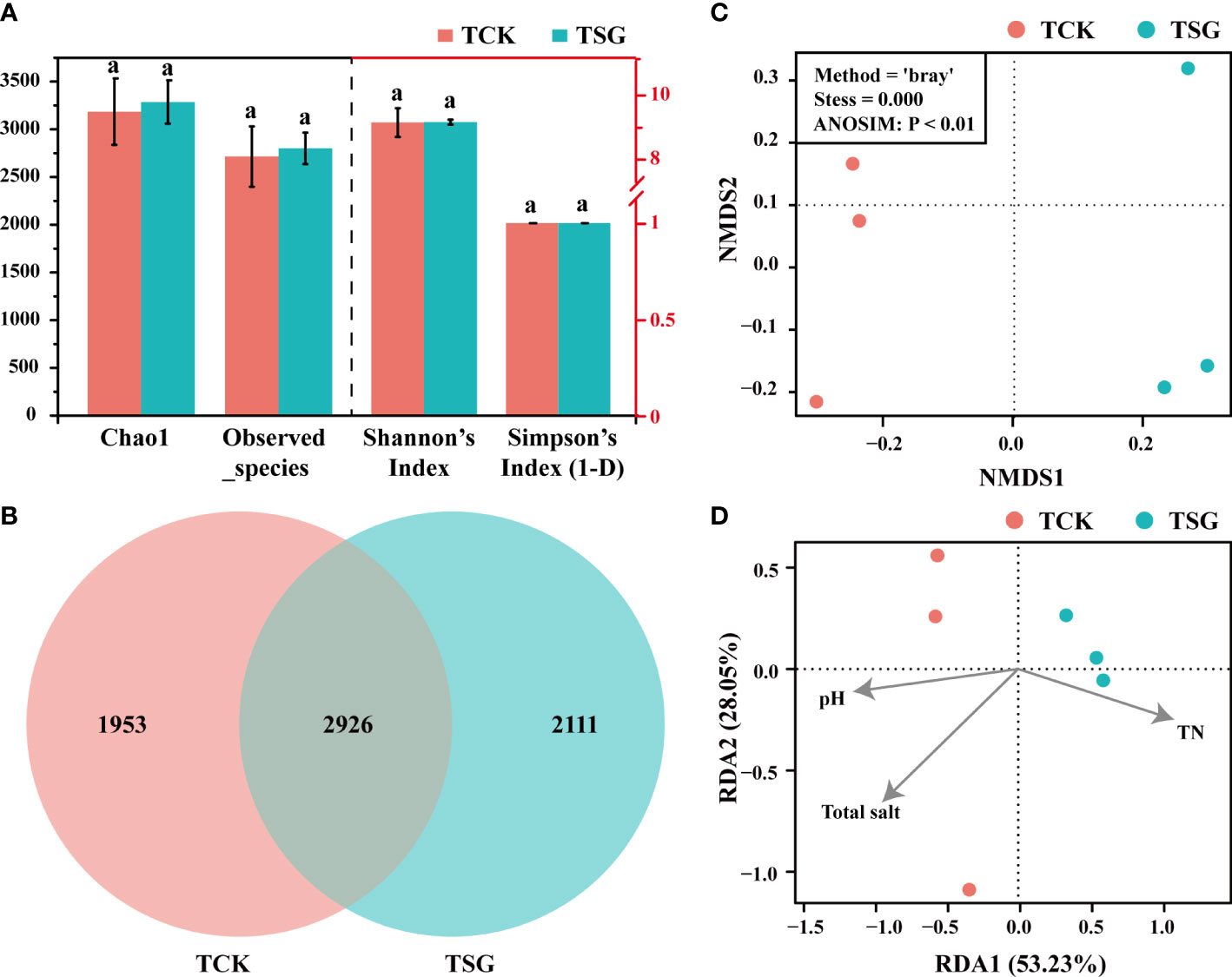
Figure 2 (A) Analysis of alpha diversity (The different lowercase letters of the same index in different sample groups indicate that there is a significant difference between the groups in this index [according to Duncan’s significance test, P < 0.05)]; (B) Venn diagram showing the number of shared/unique OTUs between two groups of samples; (C) NMDS diagram showing bacterial community differences between two sample groups; (D) RDA diagram showing the correlation between bacterial community composition and environmental factors in the two groups of samples.
Redundancy analysis (RDA) was conducted on the bacterial community composition structure and environmental factors of the two groups of samples. The soil pH, total salt, and TN with significant differences between the two groups were selected as environmental factors, and the RDA results are displayed in Figure 2D. In RDA, the variable ray is extended, the sample is projected vertically on the ray, and the value of the environmental variable increases along the direction of the variable arrow. The results indicated that the bacterial community composition of TCK was positively correlated with environmental factors related to saline-alkali stress (total salt and pH); while TSG was positively correlated with TN. Therefore, the rhizosphere bacterial community composition of S. glauca in TCSS was related to the decrease in saline-alkali stress and accumulation of nitrogen.
3.2.2. Analysis of bacterial community composition
The horizontal sequences at all levels were classified from phylum to genus, and the community composition results at dominant phylum and genus levels are illustrated in Figure 3. There were 44 phyla in the bacterial sequence. The abundance difference between the two samples (Figure 3A) of the phyla TOP15 was evaluated, and the results found that the dominant phyla (relative abundance > 1%) between the two samples were similar. Among them, Proteobacteria, as the most dominant phylum in the two samples, had an abundance exceeding 60%; the difference was that the abundance of Deltaproteobacteria in TSG was decreased. In addition, compared with TCK, the abundance of Bacteroidetes and Actinomycetes in TSG increased, while the abundances of Acidobacteria and Nitrospira decreased, which was similar to the trend of bacterial community composition in the rhizosphere of S. glauca in original CSS (An et al., 2022). The results showed that the growth of S. glauca in different habitats enriched similar bacterial communities, suggesting that the formation of specific bacterial communities was closely related to plant types.
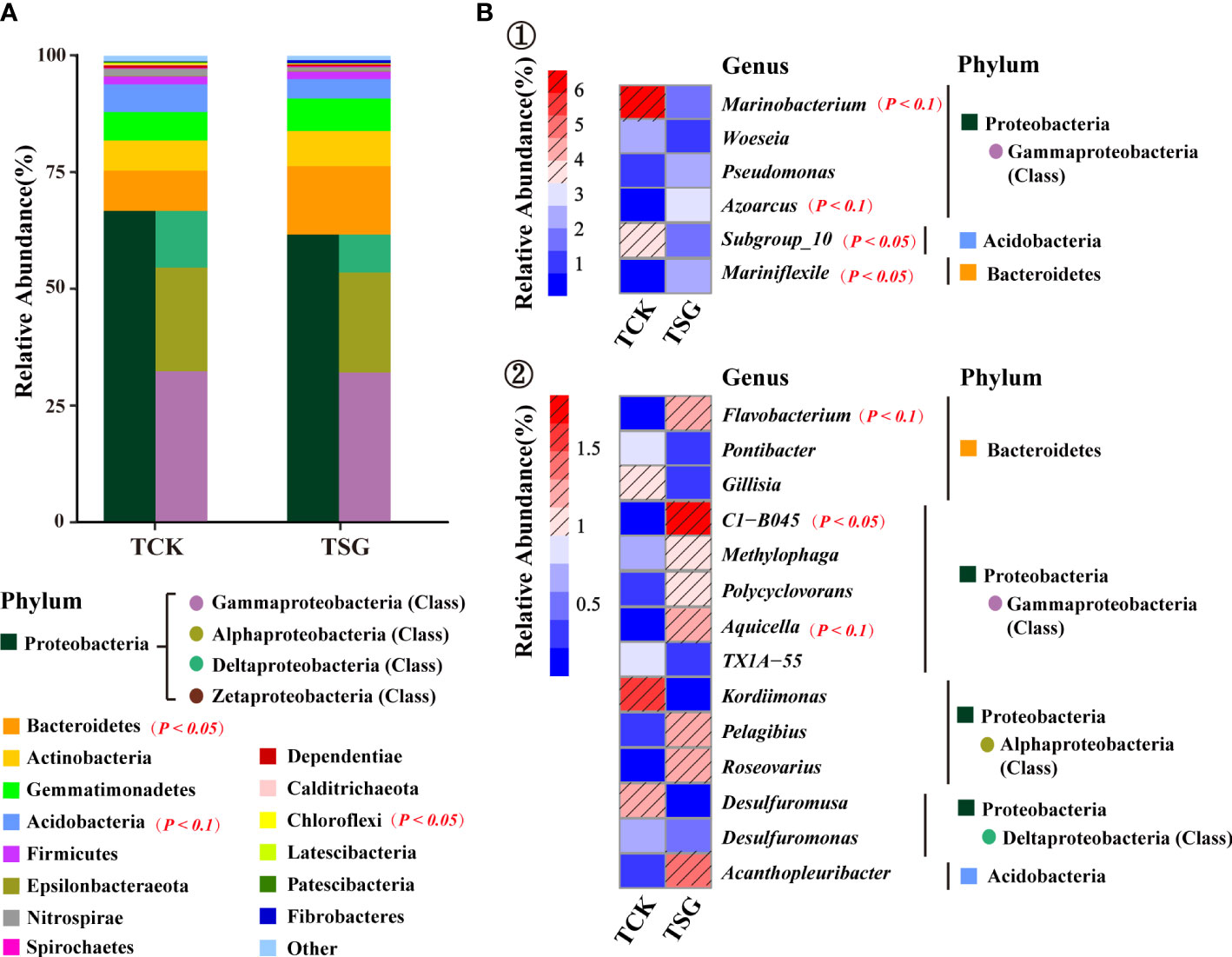
Figure 3 (A) Abundances of the dominant TOP 15 phyla in two groups of samples; (B) Abundances of the dominant TOP 20 genera in two groups of samples (① and ② use different abundance scales).
In order to more clearly describe the difference of genus-level community composition between the two groups of samples, the community composition of top-20 genera of the two groups of samples was visualized using a heat map (Figure 3B). Marinobacterium (TCK 6.56% > TSG 1.47%), Woeseia (TCK 2.26% > TSG 1.19%), and Subgroup_10 (TCK 3.49% > TSG 1.67%) were dominant genera in both groups (relative abundance > 1%). In addition, Kordiimonas (1.60%) and Desulfuromusa (1.24%) were dominant genera in TCK. The dominant genera of TSG included Gammaproteobacteria Azoarcus (2.72%), Pseudomonas (2.16%), C1-B045 (1.84%), Aquicella (1.18%), Methylophaga (1.05%); Alphaproteobacteria Roseovarius (1.20%), and Pelagibius (1.17%); Bacteroidetes Mariniflexile (2.08%), Flavobacterium (1.27%), Acidobacteria Acanthopleuribacter (1.32%), etc. The results indicated that the rhizosphere soil of S. glauca enriched more dominant bacterial populations than the bulk soil, which may have important ecological functions.
3.2.3. Analysis of bacterial species with significant differences between groups
In order to further determine the different information of bacterial communities between groups, the differential species (named Biomarkers) that contribute significantly to the differences between groups were further studied by Linear discriminant Effect Size (LEfSe) analysis. Among them, the linear discriminative analysis (LDA) score was adopted to estimate the contribution of different species to the difference between groups. The bigger the absolute value of the LDA value, the greater the influence of the species as a significantly different species on the difference in bacterial diversity between groups. In this study, the LEfSe analysis histogram (Figure 4) demonstrated significantly different bacterial taxa (P< 0.05) with an absolute value of LDA score exceeding 4.
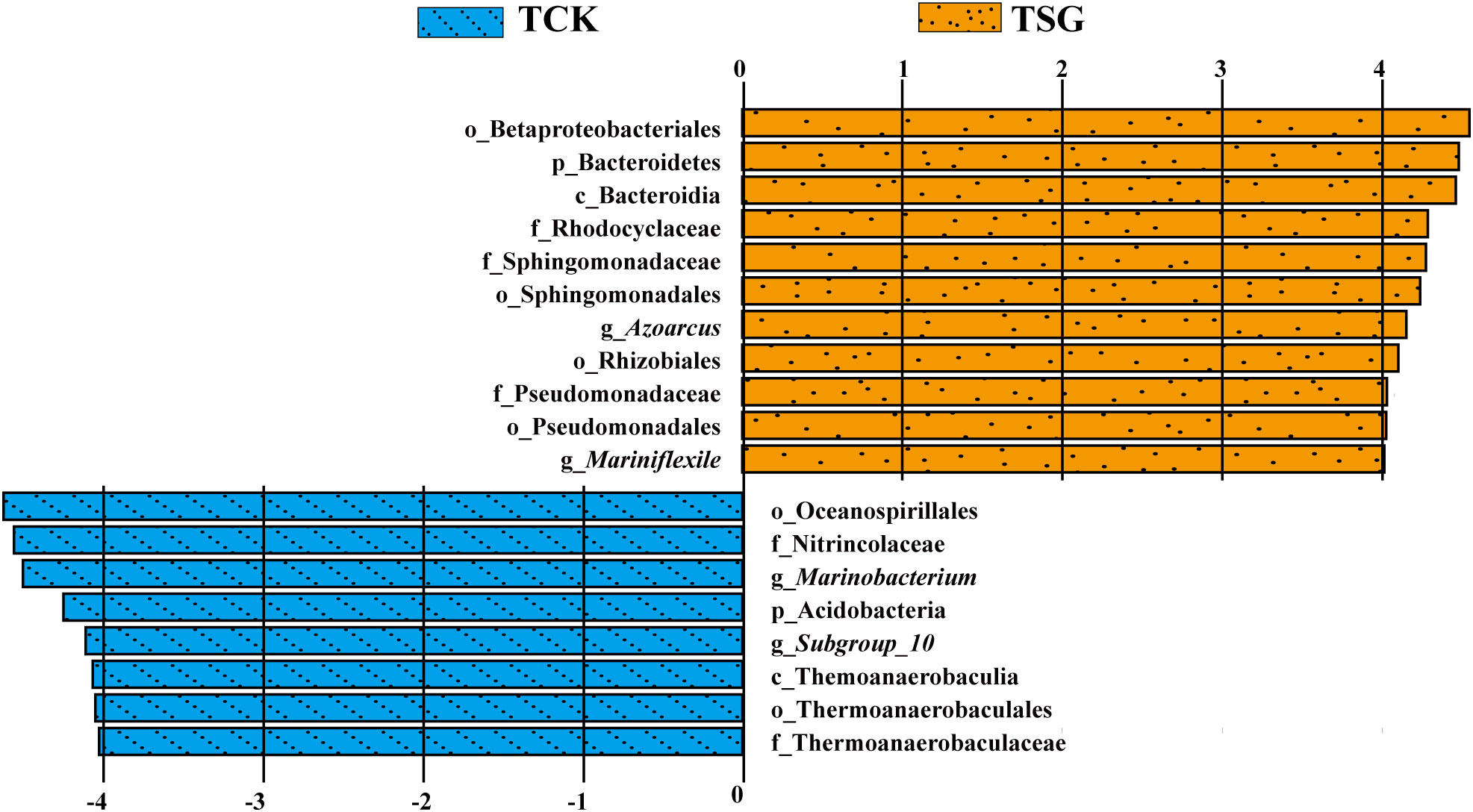
Figure 4 Column diagram of LEfSe analysis for two groups of samples (“p” denotes phylum-level; “c” denotes class-level; “o” denotes order-level; “f” denotes family-level; “g” denotes genus-level; the length of the columns represents the effect size of the significantly different species, which is plotted according to the LDA score (log_10)).
As shown in Figure 4, only in the smallest taxonomic rank, the biomarkers in TCK include g_Marinobacterium (belonging to f_Nitrincolaceae, o_Oceanospirillales) and g_Subgroup_10 (belonging to f_Thermoanaerobaculaceae, o_Thermoanaerobaculales, c_Thermoanaerobaculia, and p_Acidobacteria). Biomarkers in TSG consists of g_Azoarcus (belonging to f_Rhodocyclaceae, o_Betaproteobacteriales), g_Mariniflexile (belonging to c_Bacteroidia, p_Bacteroidetes), f_Sphingomonadaceae (belonging to o_Sphingomonadales), o_Rhizobiales, and f_Pseudomonadaceae (belonging to o_Pseudomonadales). The results showed that there were more biomarkers in the rhizosphere of S. glauca than those in bulk soil, and these biomarkers may play a vital role in the bacterial community structure of the corresponding groupings.
3.2.4. Co-occurrence network analysis of bacterial communities
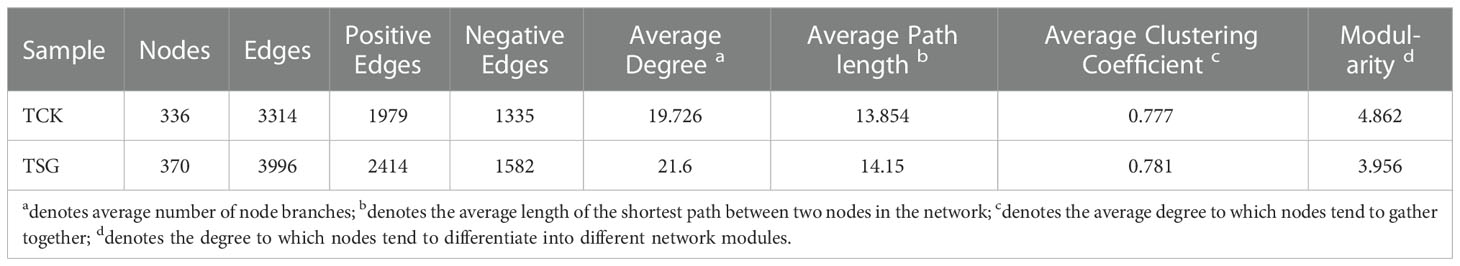
The bacterial community co-occurrence network of the two groups of samples was established at the genus level (Figure 5). Compared with TCK, the number of nodes, number of edges, average degree, average path length, and other indices of bacterial communities in TSG were higher (Table 2). The finding indicated that the bacterial co-occurrence network structure in rhizosphere of S. glauca was more complex. On the contrary, the modular level of rhizosphere bacterial groups in S. glauca was low (Table 2), indicating that the network modules of rhizosphere bacterial groups in S. glauca were relatively dispersed. Moreover, central nodes with high degree (> 30) and tight centrality (> 0.15) in collinear networks represent different bacterial genera (Tables S2, S3). Obviously, the co-occurrence network structure of the bacterial community in the rhizosphere of S. glauca and the bulk soil differs, while the co-occurrence relationship of the bacterial community in the rhizosphere of S. glauca is more complex.
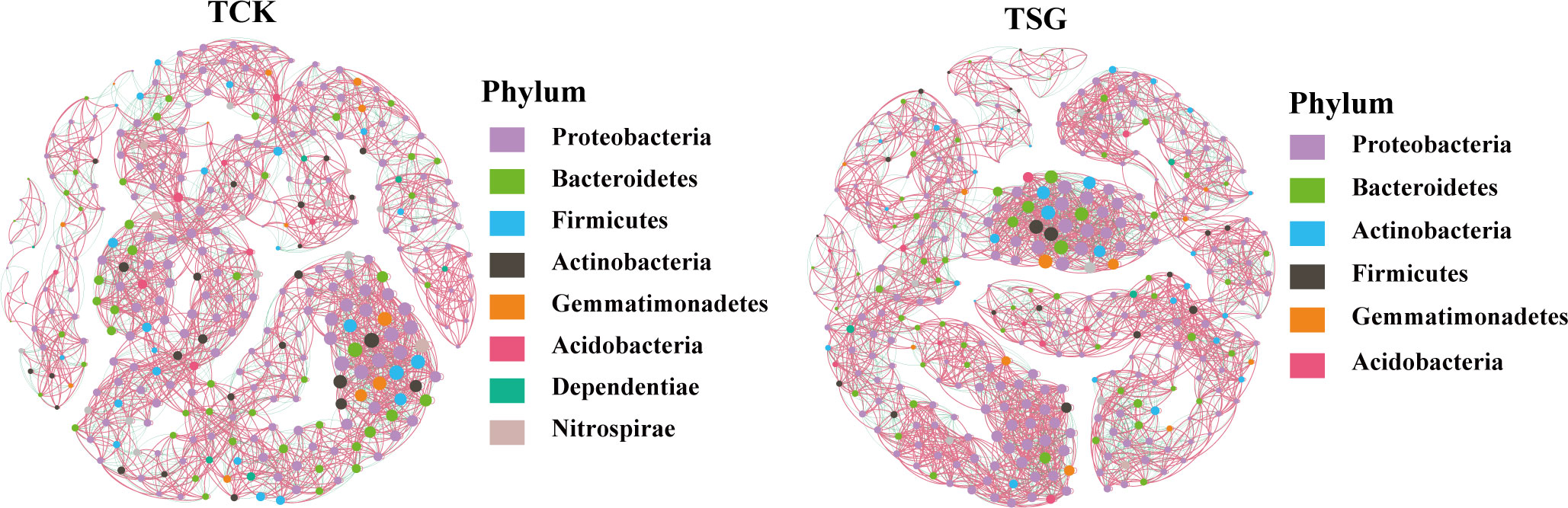
Figure 5 Co-occurrence networks between bacterial taxa based on genus at the Phylum level according to the Pearson correlation coefficients in sample TCK and TSG (Nodes labeled with the same color indicate that different genus belonged to the same bacterial Phylum; node size indicates link numbers with other nodes and larger sizes indicate more link numbers; the edge represents the connected line between two nodes with significant correlation (Pearson’s correlation coefficient > 0.6, P< 0.05); the red and green lines within the two nodes in the networks represent positive and negative correlations, respectively.).
3.2.5. Predictive analysis of bacterial community function
The FAPROTAX database can be used to annotate prokaryotic microbial metabolism or other ecologically related functions (e.g. nitrification, denitrification, and fermentation) (Louca et al., 2016). In this study, the bacterial community functions of two groups of samples were predicted based on the FAPROTAX database. The results of functional prediction of the top-30 are shown in Figure 6. The chemoheterotrophy and aerobic_chemoheterotrophy functions of bacterial communities in the two groups of samples were dominant; in addition, fermentation, N metabolism related (e.g. nitrate_reduction, nitrate_respiration, nitrogen_respiration, and nitrite_respiration), pathogen related, and phototrophy related (e.g. phototrophy, photoheterotrophy) were dominant in both groups of samples (relative abundance > 1%). The difference is that compared with TCK, the functional abundance of aerobic_chemoheterotrophy in the rhizosphere bacterial community of S. glauca was reduced significantly, while the functional abundance of phototrophy (photoheterotrophy) was increased by more than 1%; at the same time, the functional abundance of N metabolism (nitrate_reduction, nitrate_respiration, nitrogen_respiration) and methylotrophy-related (methylotrophy, methanol_oxidation) in the rhizosphere bacterial community of S. glauca increased (Table S4). These results implied that the functions related to N metabolism and C cycling (phototrophic and methylotrophic) of the rhizosphere bacterial community of S. glauca were activated to differing extents, which could accelerate soil material circulation.
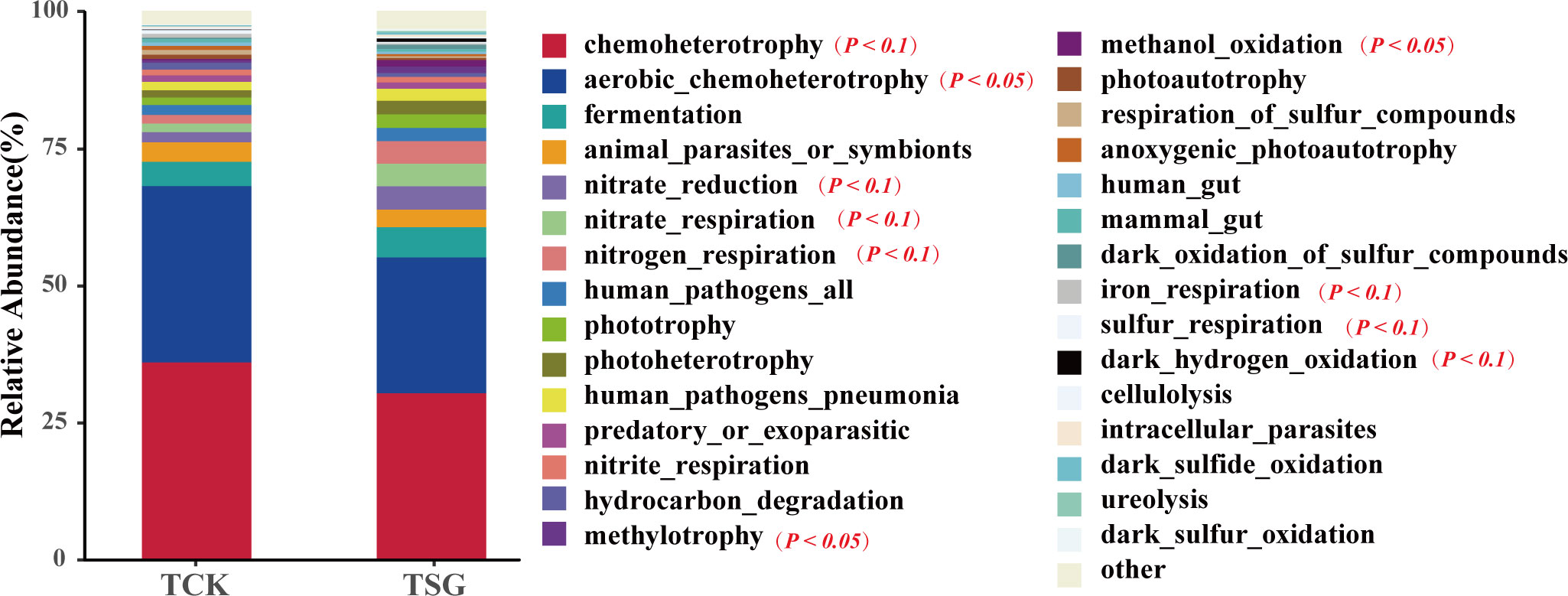
Figure 6 Relative abundance histogram of TOP-30 bacterial community function prediction based on FAPROTAX for two groups of samples.
4. Discussion
4.1. Characteristics of bacterial community in rhizosphere soil of S. glauca and its driving effect on carbon and nitrogen cycle
4.1.1. Characteristics and action process of rhizosphere-associated carbon cycle bacterial communities in S. glauca
Carbon is the element with the largest demand for microbial cells. The dominant bacteria involved in the carbon cycle in bulk soil (TCSS) include organic matter degrading bacteria such as Marinobacterium (Koshlaf et al., 2020) and Kordiimonas (Zhang et al., 2016; Girard et al., 2020) (Figures 3B, 4). Among them, Marinobacterium is a bacterium that is enriched in the bulk soil and has the highest abundance at genus level, as a type of moderately halophilic bacteria, it is highly dependent on salinity (Koshlaf et al., 2020). In other words, the bulk soil without S. glauca grows more halophilic organic matter degrading bacteria. Salt is the main factor affecting the composition of TCSS bulk soil bacterial community structure (Figure 2D), which distinctly affects the biodiversity of organic matter degrading bacteria.
The difference is that, in the TCSS environment with S. glauca growing, plant root exudates produced by plants lead to greater enrichment of organic matter degrading bacteria (Lai et al., 2011; Zhang et al., 2020). In addition, the salt content of the soil around the roots of the S. glauca was significantly reduced (Table 1), which was also conducive to the increase in the types of organic matter-degrading bacteria with dominant abundance in the rhizosphere (Figure 3), including Bacteroidetes (Mariniflexile, Flavobacterium), some Gammaproteobacteria (e.g. C1-B045), and Actinobacteria (Cockell et al., 2009; Puttaswamygowda et al., 2019). Among them, Mariniflexile is more enriched in the rhizosphere samples of S. glauca (Figure 4). An et al. (2022) explored the rhizosphere bacterial communities of three dominant salt-tolerant plants S. glauca, Tripolium vulgare, and Phragmites australis in CSS. Their results found that Mariniflexile was the dominant abundance of bacteria enriched only in the rhizosphere of S. glauca. These organic substances degrade bacteria, mainly Mariniflexile, and when enriched in the rhizosphere of S. glauca, can activate the soil C cycle and strengthen the utilization of soil organic matter, thus laying a material foundation for the enrichment of more functional bacteria in the rhizosphere of S. glauca.
In addition to the enrichment of organic matter degrading bacteria, methyl trophic bacteria and photosynthetic bacteria with carbon activation and carbon capture functions were enriched (An et al., 2022), which played an essential role in soil carbon cycle and energy metabolism, including C1 compounds (e.g. methanol) and CO2 fixation (Kolb, 2009; Hunger et al., 2011). The results of bacterial community functional prediction indicated that the bacterial functions of phototrophic and methylotrophic were also improved in rhizosphere of S. glauca (Figures 6, S4). Among them, the rhizosphere of S. glauca was significantly enriched in Methylophaga (Figure 3B), which could conduct carbon assimilation with C1 compound as the sole growth substrate (Boden, 2012), realizing carbon emission reduction and increase in carbon-sink capacity. In addition, the rhizosphere of S. glauca was significantly enriched with a variety of photosynthetic bacteria, including the dominant bacterium Roseovarius (1.20%), and the dominant differential species f_Sphingomonadaceae (containing high abundances of Erythrobacter 0.77%) (Figures 3B, 4). Roseovarius and Erythrobacter contain bacterial chlorophyll A, which can grow in aerobic, but no oxygen production, and confers photosynthetic functions (Jiao et al., 2003; Jiao, 2012).
4.1.2. Characteristics and action process of rhizosphere-associated nitrogen cycle bacterial communities in S. glauca
The nitrogen cycle is an important part of the material cycle of coastal ecosystem, which is of great significance for maintaining coastal ecological balance and promoting coastal restoration (Kuypers et al., 2018). The content of TN in the bulk soil of TCSS was poor, while the content of TN in the soil around the root of S. glauca was significantly increased (Table 1). The TN content was the main factor influencing bacterial community composition in rhizosphere of S. glauca (Figure 2D), and functional bacteria involved in nitrogen cycling were significantly enriched in the rhizosphere of S. glauca (Figures 3B, 4), including: Azoarcus with nitrogen fixation (Raittz et al., 2021), and Pseudomonas with both denitrification and dissimilatory nitrate reduction to ammonium (DNRA) pathways (Su et al., 2012; Huang et al., 2020b). Meanwhile, the abundance of the nitrifying bacteria Nitrospirae in the rhizosphere of S. glauca decreased, but still dominated the bacterial community (TOP 7, 0.83%) (Figure 3A). Nitrogen fixation, nitrification, and denitrification are the most important processes in the nitrogen cycle (Seitzinger, 1988; Laverman et al., 2007; Wei, 2012). The rhizosphere of S. glauca grown in TCSS enriched the N-cycle functional bacteria which were significantly different from that grown in bulk soil, which helped to achieve soil nitrogen accumulation and was beneficial to plant growth and soil fertility improvement.
In addition to being beneficial to nitrogen accumulation, the nitrogen cycle activated by the rhizosphere bacterial community of S. glauca also helps maintain soil biogeochemical cycles and drives coastal material transformation and energy metabolism. In this study, the nitrogen respiration-related functions of the rhizosphere bacterial community of S. glauca were significantly enhanced (Figures 6, S4). The reduction reaction of nitrogen respiration and the oxidative reaction of organic matter degradation under hypoxic conditions were coupled by electron transport (Boyd, 1995; Bulseco et al., 2019), which jointly promoted the biogeochemical cycle in the soil. In addition, the content in TCSS was relatively low due to the cover and modification of sea-sand (Table 1). In addition to the leaching effect, bacterial groups characterized by the reducing function of sulfur-containing compounds were enriched in the bulk soil of TCSS without plant growth (Figures 3, 4), including: Subgroup_10, which can reduce elemental sulfur to hydrogen sulfide (Izumi et al., 2012), and sulfate-reducing bacteria Deltaproteobacteria (including Desulfuromonas and Desulfuromusa), etc. These bacteria can use the electrons generated during the degradation of organic matter to reduce sulfur-containing compounds, which is akin to the action of rhizosphere bacteria in S. glauca growing in CSS (An et al., 2022). However, TCSS has a high porosity (Table 1), and the growth of S. glauca in TCSS further creates an oxygen-rich environment in the rhizosphere. This results in the growth of strictly anaerobic sulfur-reducing bacteria (Vandieken et al., 2006; Flieder et al., 2021) did not dominate in the rhizosphere of S. glauca (Figure 3A). In conclusion, S. glauca grows in TCSS, and the life process of its rhizosphere bacteria has transformed from the sulfur cycle in bulk soil to the nitrogen cycle, which is conducive to the accumulation and utilization of nitrogen in the soil and drives the benign transformation of soil structure.
4.2. Pathway of S. glauca growth promoting benign development of TCSS soils
The CSS accumulated through the reclamation project is silty soil under severe salt stress, with high water content and poor permeability (Table 1), while the large-scale CSS was modified by sea-sand cover (TCSS) in 2015. The porosity of TCSS after modification is relatively high, which indicates that the TCSS leaching ability is stronger under the same precipitation conditions; the soil salinity is relatively low, which indicates that the TCSS is under mild or severe salt stress, but the total calcium content of TCSS exceeds 1%, showing the characteristics of calcareous saline-alkali soil (Bao, 2000; Lv & Li, 2010); the contents of nutrients such as N, P and the ability of soil to retain fertilizer in TCSS are relatively low (Table 1), according to the second national soil survey and relevant standards (National Soil Survey Office, 1998), the soil nutrients here are classified as poor. Further, Pearson correlation analysis results between soil physico-chemical properties (Table S1) show that soil salt content, nutrient content, and fertilizer retention performance are significantly negatively correlated with soil leaching performance. Affected by the improvement of soil leaching performance, the soil salt stress pressure of TCSS is relatively small, but there are disadvantages of soil nutrient leaching, poor soil, and poor fertilizer retention performance, which cause many kinds of plants to fail to grow well.
The plant S. glauca is a polysalt-type halophyte, which can accumulate salt in the body and reduce soil salinity (Wang et al., 2004; Han et al., 2005; Mori et al., 2011). The field investigation in this area shows that wild S. glauca is the only species that can form a single vegetative community in TCSS (Figure 1C), and there is more accumulation of salt-based ions in the aboveground parts of wild S. glauca (Figure S1). Therefore, the growth of S. glauca reduces salt stress in TCSS. In the meantime, the total nitrogen content of the soil around the roots of S. glauca grown by TCSS significantly increased (Table 1), which is closely associated with the enrichment of bacterial communities related to nitrogen fixation (Figures 3B, 4) and the nitrogen cycle (Figures 6, S4) in the rhizosphere of S. glauca, laying a foundation for nitrogen accumulation in TCSS. Based on this, S. glauca grows in TCSS, which reduces soil salinity and increases total nitrogen content, enriches and constructs a beneficial bacterial community structure. This can promote the benign development of soil, thereby promoting soil improvement and soil fertility.
The bacterial richness and evenness in the rhizosphere of S. glauca in TCSS are akin to that in bulk soil (Figure 2A), but the composition of bacterial community structure is significantly different from that in bulk soil (Figures 2B, C, 3). At the same time, the co-occurrence relationship of bacterial community in the rhizosphere of S. glauca is more complex, and the interconnected network is denser (Figure 5), indicating that its rhizosphere soil may have a more refined metabolic regulation relationship. Many studies have shown that the life activities of plant rhizosphere microorganisms exert an important influence on improving soil properties and promoting the benign development of the plant-microbe symbiosis system (Gutknecht et al., 2006; Mendes et al., 2013; Lynch, 2019; Bhuyan et al., 2020; Trivedi et al., 2020). Based on the above analysis of the key material cycle process of the rhizosphere dominant bacterial community participating of S. glauca in TCSS, the predictive path of action is proposed such that the bacterial community in rhizosphere of S. glauca promotes the benign development of TCSS soil by enriching and enhancing the C/N cycles (Figure 7).
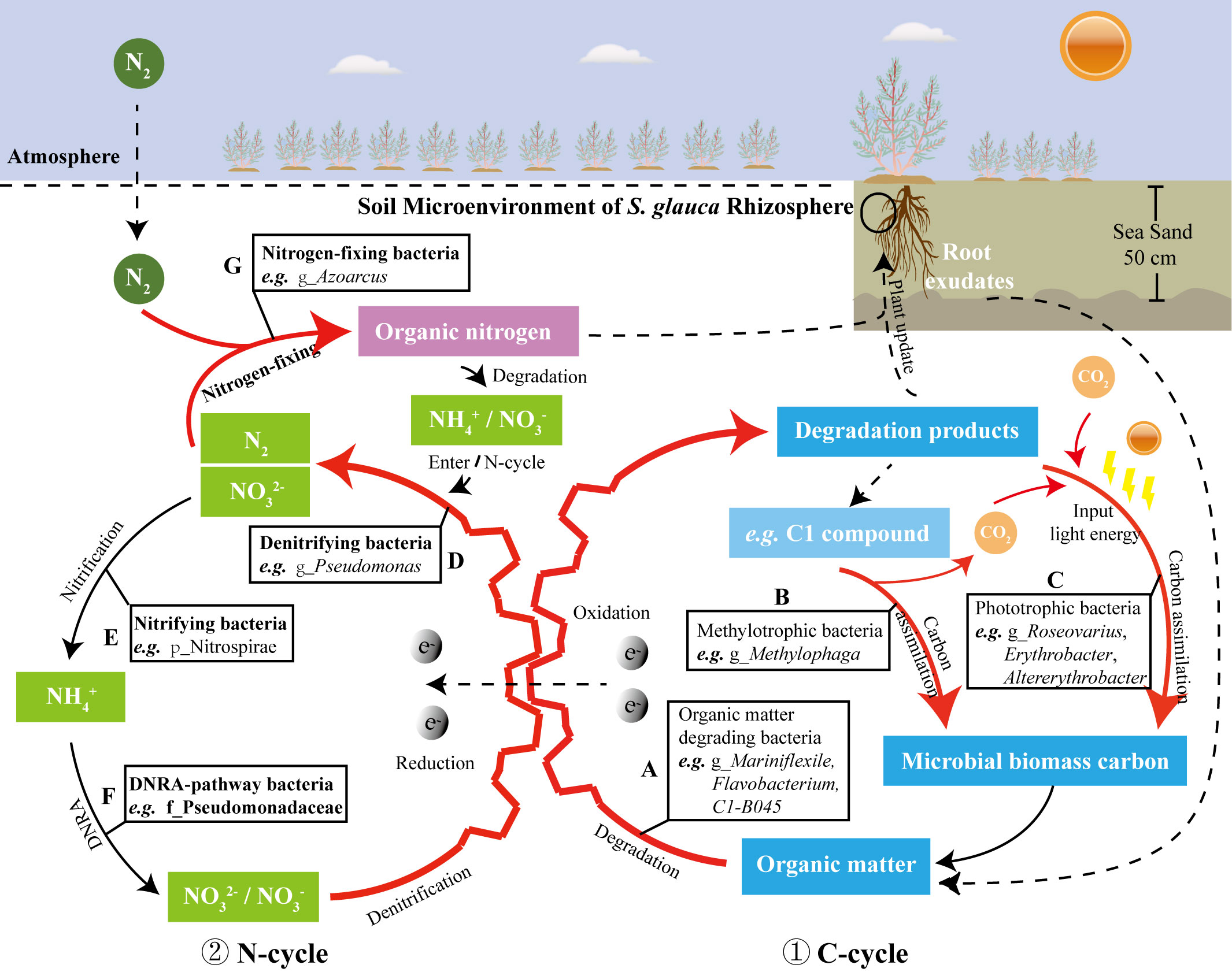
Figure 7 The dominant bacterial community in the rhizosphere of S. glauca were predicted to participates in the C/N cycle of TCSS (the red bold lines represent the C/N cycle pathway with enhanced enrichment of the rhizosphere bacterial community of S. glauca); The serial number ① indicates the predicted C-cycle model; the serial number ② indicates the predicted N-cycle model; the different capital letters in (A–G) refer to the functional bacteria involved in different stages of the C/N cycle, respectively.
As shown in Figure 7, various dominant bacteria enriched in the rhizosphere of S. glauca are mainly related to the C/N cycle. The bacteria related to the carbon cycle (Figure 7, ①) mainly include organic matter degrading bacteria, methyl trophic bacteria, and phototrophic bacteria, etc. Many different types of organic matter that can degrade bacteria (Figure 7A) are enriched with the rhizosphere exudates of S. glauca and the refractory organic matter in the soil as the substrate. These organic matter degrading bacteria generate a variety of degradation products for the growth of plants and heterotrophic microorganisms. Among them, the dominant heterotrophic methylotrophic bacteria (Figure 7B) use the secondary metabolite C1 compound as the sole growth substrate, which is converted into the components of the organism or generate CO2 through the carbon assimilation process; carbon-capturing bacteria (phototrophic-bacteria) (Figure 7C) use light energy to synthesize a significant amount of microbial carbon, which is an important component of soil organic carbon. The bacteria related to the nitrogen cycle (Figure 7, ②) have the functions (see Section 4.2) of nitrogen fixation (Figure 7G), denitrification (Figure 7D), nitrification (Figure 7E), and dissimilatory reduction of nitrate to ammonium (Figure 7F). Among them, the bacterial community is dominated by nitrogen respiration (Figure 6). The substrate nitrate/nitrite of nitrogen respiration is the electron receptor of organic matter degradation reaction (oxidation reaction) under the condition of soil hypoxia (Figures 7A, D) and participated in the reaction, therefore, the enhancement of nitrogen respiration coupled organic matter degradation and promote the circulation and transformation of soil substances.
To be clear, the detection of specific microbes (or genes) in soil does not imply activity of these microbes – or else many microbes can be present but functioning in a different way given how versatile many taxa are. Combined with existing literature reports and based on the analysis of soil physicochemical properties, a reasonable analysis of the soil biogeochemical cycle in which these bacterial communities may participate is helpful to increase the understanding of the function of the rhizosphere bacterial community in the soil samples, thus establishing a solid foundation for the subsequent microbial application research.
5. Conclusion
In summary, the CSS modified by sea-sand is calcareous saline-alkali soil, and wild S. glauca growing in this soil can significantly reduce soil saline-alkali stress and increase the nitrogen content of the soil. The positive correlation with the benign changes of soil environment is that the functions of nitrogen respiration, photo-nutrition, and methyl nutrition of the rhizosphere bacterial community of S. glauca are strengthened, and the co-occurrence relationship of the bacterial community tends to be complicated. Meanwhile, the rhizosphere of S. glauca is significantly enriched with a variety of functional bacteria involved in the C and N cycles, while these functional bacteria were predicted to promote soil carbon activation and carbon capture, and also increase the effect of soil nitrogen fixation, so that making TCSS further improved. In a word, large-scale cultivation of S. glauca or combination of S. glauca and other suitable plants in the Lianyungang coastal zone is conducive to the construction of soil bacterial community that promotes the benign development of soil, thus improving soil fertility and realizing the precise improvement of TCSS. The results provide a scientific basis for the local government to conduct coastal ecological restoration, and offer a model for the improvement of similar coastal saline-alkali land around the world.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA792088.
Author contributions
BL conceived and supervised the study, XA conducted experiments, analyzed the data and wrote original draft. BL, XA and XT carried out investigation on the test site. BL, ZW, KJ, XT, RZ, and MX participanted in the discssion and revisions of the manuscript. MX, ZW and BL: Funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was jointly supported by the Jiangsu Provincial Marine Science and Technology Innovation Special Project [grant number HY2019-3]; the Graduate Research and Innovation Projects of Jiangsu Province [grant number KYCX21_1360]; the Interdisciplinary Project of Nanjing Normal University [grant number 164320H1847].
Acknowledgments
The author would like to thank Mr. Wang Xiao, Ms. Yanshuang Liu, and Mr. Jin Xu of Lianyungang Golden Coast Development and Construction Co. LTD (Lianyungang City, Jiangsu Province, China) for their support in the site investigation process.
Conflict of interest
Authors XT and RZ were employed by Lianyungang Golden Coast Development and Construction Co. LTD.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1001449/full#supplementary-material
References
An X. C., Wang Z. F., Teng X. M., Zhou R. R., Wang X. X., Xu M., et al. (2022). Rhizosphere bacterial diversity and environmental function prediction of wild salt-tolerant plants in coastal silt soil. Ecol. Indic. 134, e108503. doi: 10.1016/j.ecolind.2021.108503
Ariel A., Feitelson E., Marinov U. (2021). Economic and environmental explanations for the scale and scope of coastal management around the Mediterranean. Ocean. Coast. Manage. 209, e105639. doi: 10.1016/j.ocecoaman.2021.105639
Belle-Oudry D. (2008). Quantitative analysis of sulfate in water by indirect EDTA titration. J. Chem. Educ. 85, e1269. doi: 10.1021/ed085p1269
Bhuyan B., Debnath S., Pandey P. (2020). “The rhizosphere microbiome and its role in plant growth in stressed conditions,” in Rhizosphere microbes. microorganisms for sustainability. Eds. Sharma S. K., Singh U. B., Sahu P. K., Singh H. V., Sharma P. K. (Singapore: Springer), 503–509. doi: 10.1007/978-981-15-9154-9
Boden R. (2012). Emended description of the genus Methylophaga janvier et al. 1985. Int. J. Syst. Evol. Microbiol. 62 (7), 1644–1646. doi: 10.1099/ijs.0.033639-0
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Bouchez T., Blieux A. L., Dequiedt S., Domaizon I., Dufresne A., Ferreira S., et al. (2016). Molecular microbiology methods for environmental diagnosis. Environ. Chem. Lett. 14 (4), 423–441. doi: 10.1007/s10311-016-0581-3
Boyd C. E. (1995). “Soil organic matter, anaerobic respiration, and oxidation-reduction,” in Bottom soils, sediment, and pond aquaculture. Ed. Boyd C. E. (New York, NY: Springer), 194–218. doi: 10.1007/978-1-4615-1785-6
Bulseco A. N., Giblin A. E., Tucker J., Murphy A. E., Sanderman J., Hiller-Bittrolff K., et al. (2019). Nitrate addition stimulates microbial decomposition of organic matter in salt marsh sediments. Glob. Change Biol. 25 (10), 3224–3241. doi: 10.1111/gcb.14726
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 (5), 335–336. doi: 10.1038/nmeth.f.303
Chi Z. F., Wang W. J., Li H., Wu H. T., Yan B. X. (2021). Soil organic matter and salinity as critical factors affecting the bacterial community and function of Phragmites australis dominated riparian and coastal wetlands. Sci. Total Environ. 762, e143156. doi: 10.1016/j.scitotenv.2020.143156
Cockell C. S., Olsson K., Knowles F., Kelly L., Herrera A., Thorsteinsson T., et al. (2009). Bacteria in weathered basaltic glass, Iceland. Geomicrobiol J. 26 (7), 491–507. doi: 10.1080/01490450903061101
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 (16), 2194–2200. doi: 10.1093/bioinformatics/btr381
Flieder M., Buongiorno J., Herbold C. W., Hausmann B., Rattei T., Lloyd K. G., et al. (2021). Novel taxa of acidobacteriota implicated in seafloor sulfur cycling. ISME J. 15 (11), 3159–3180. doi: 10.1038/s41396-021-00992-0
Girard E. B., Kaliwoda M., Schmahl W. W., Wörheide G., Orsi W. D. (2020). Biodegradation of textile waste by marine bacterial communities enhanced by light. Env. Microbiol. Rep. 12 (4), 406–418. doi: 10.1111/1758-2229.12856
Gómez C., White J. C., Wulder M. A. (2016). Optical remotely sensed time series data for land cover classification: a review. ISPRS J. Photogramm. 116, 55–72. doi: 10.1016/j.isprsjprs.2016.03.008
Gutknecht J. L. M., Goodman R. M., Balser T. C. (2006). Linking soilprocess and microbial ecology in freshwater wetland ecosystems. Plant Soil. 289, 17–34. doi: 10.1007/s11104-006-9105-4
Han N., Shao Q., Lu C. M., Wang B. S. (2005). The leaf tonoplast V-H+-ATPase activity of a C3 halophyte Suaeda salsa is enhanced by salt stress in a Ca-dependent mode. J. Plant Physiol. 162 (3), 267–274. doi: 10.1016/j.jplph.2004.07.016
He C., Zheng L., Ding J., Gao W., Li Q., Han B., et al. (2022). Variation in bacterial community structures and functions as indicators of response to the restoration of Suaeda salsa: A case study of the restoration in the beidaihe coastal wetland. Front. Microbiol. 13, e783155. doi: 10.3389/fmicb.2022.783155
Huang L. B., Bai J. H., Wen X. J., Zhang G. L., Zhang C. D., Cui B. S., et al. (2020a). Microbial resistance and resilience in response to environmental changes under the higher intensity of human activities than global average level. Glob Change Biol. 26 (4), 2377–2389. doi: 10.1111/gcb.14995
Huang X., Weisener C. G., Ni J., He B., Xie D., Li Z. (2020b). Nitrate assimilation, dissimilatory nitrate reduction to ammonium, and denitrification coexist in Pseudomonas putida y-9 under aerobic conditions. Bioresour Technol. 312, e123597. doi: 10.1016/j.biortech.2020.123597
Hunger S., Schmidt O., Hilgarth M., Horn M. A., Kolb S., Conrad R., et al. (2011). Competing formate- and carbon dioxide-utilizing prokaryotes in an anoxic methane-emitting fen soil. Appl. Env. Microbiol. 77 (11), 3773–3785. doi: 10.1128/AEM.00282-11
Izumi H., Nunoura T., Miyazaki M., Mino S., Toki T., Takai K., et al. (2012). Thermotomaculum hydrothermale gen. nov. sp. nov. a novel heterotrophic thermophile within the phylum acidobacteria from a deep-sea hydrothermal vent chimney in the southern Okinawa trough. Extremophiles 16 (2), 245–253. doi: 10.1007/s00792-011-0425-9
Jiang T. T., Pan J. F., Pu X. M., Wang B., Pan J. J. (2015). Current status of coastal wetlands in China: degradation, restoration, and future management. Estuar. Coast. Shelf S. 164, 265–275. doi: 10.1016/j.ecss.2015.07.046
Jiao N. Z. (2012). Carbon fixation and sequestration in the ocean, with special reference to the microbial carbon pump. Scientia Sin. (Terrae). 42 (10), 1473–1486. Available at: https://www.researchgate.net/publication/284886634.
Jiao N. Z., Sieracki M. E., Zhang Y., Du H. L. (2003). Aerobic anoxygenic phototrophic bacteria and their roles in marine ecosystems. Chin. Sci. Bull. 48 (6), 530–534. doi: 10.1360/02wc0336
Kolb S. (2009). Aerobic methanol-oxidizing bacteria in soil. FEMS Microbiol. Lett. 300 (1), 1–10. doi: 10.1111/j.1574-6968.2009.01681.x
Koshlaf E., Shahsavari E., Haleyur N., Osborn A. M., Ball A. S. (2020). Impact of necrophytoremediation on petroleum hydrocarbon degradation, ecotoxicity and soil bacterial community composition in diesel-contaminated soil. Environ. Sci. pollut. Res. 27 (25), 31171–31183. doi: 10.1007/s11356-020-09339-2
Kuypers M. M. M., Marchant H. K., Kartal B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16 (5), 263–276. doi: 10.1038/nrmicro.2018.9
Lai W. L., Wang S. Q., Peng C. L., Chen Z. H. (2011). Root features related to plant growth and nutrient removal of 35 wetland plants. Water Res. 45 (13), 3941–3950. doi: 10.1016/j.watres.2011.05.002
Laverman A. M., Canavan R. W., Slomp C. P., Van Cappellen P. (2007). Potential nitrate removal in a coastal freshwater sediment (Haringvliet lake, the Netherlands) and response to salinization. Water Res. 41 (14), 3061–3068. doi: 10.1016/j.watres.2007.04.002
Lin X. Z., Shen J. H., Liu K. Z., Huang X. H. (2005). Study on remediation effects of Suaeda salsa l. planting on coastal saline soil. Adv. Mar. Biol. 23 (1), 65–69. Available at: https://www.researchgate.net/publication/284601023.
Liu F., Mo X., Kong W., Song Y. (2020). Soil bacterial diversity, structure, and function of Suaeda salsa in rhizosphere and non-rhizosphere soils in various habitats in the yellow river delta, China. Sci. Total Environ. 740, e140144. doi: 10.1016/j.scitotenv.2020.140144
Li X., Yang W., Li S., Sun T., Bai J. H., Pei J., et al. (2020). Asymmetric responses of spatial variation of different communities to a salinity gradient in coastal wetlands. Mar. Environ. Res. 158, e105008. doi: 10.1016/j.marenvres.2020.105008
Louca S., Parfrey L. W., Doebeli M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353 (6305), 1272–1277. doi: 10.1126/science.aaf4507
Lynch J. P. (2019). Root phenotypes for improved nutrient capture: an underexploited opportunity for global agriculture. New. Phytol. 223 (2), 548–564. doi: 10.1111/nph.15738
Magoč T., Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 (21), 2957–2963. doi: 10.1093/bioinformatics/btr507.i
Malathi P., Stalin P. (2018). Evaluation of AB-DTPA extractant for multinutrients extraction in soils. Int. J. Curr. Microbiol. App Sci. 7 (3), 1192–1205. doi: 10.20546/ijcmas.2018.703.141
Mendes R., Garbeva P., Raaijmakers J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37 (5), 634–663. doi: 10.1111/1574-6976.12028
Mori S., Suzuki K., Oda R., Higuchi K., Maeda Y., Yoshiba M., et al. (2011). Characteristics of na+ and k+ absorption in Suaeda salsa (L.) pall. Soil Sci. Plant Nutr. 57 (3), 377–386. doi: 10.1080/00380768.2011.586322
Murray N. J., Phinn S. R., DeWitt M., Ferrari R., Johnston R., Lyons M. B., et al. (2019). The global distribution and trajectory of tidal flats. Nature 565 (7738), 222–225. doi: 10.1038/s41586-018-0805-8
Nossa C. W., Oberdorf W. E., Yang L. Y., Aas J. A., Paster B. J., DeSantis T. Z., et al. (2010). Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 16 (33), 4135–4144. doi: 10.3748/wjg.v16.i33.4135
Olsen S. R., Cole C. V., Watanabe F. S., Dean L. A. (1954). “Estimation of available phosphorus in soils by extraction with sodium bicarbonate,” in USDA Circular no. 939 (Washington: US Government Printing Office).
Puttaswamygowda G. H., Olakkaran S., Antony A., Purayil A. K. (2019). “Present status and future perspectives of marine actinobacterial metabolites,” in Recent developments in applied microbiology and biochemistry. Ed. Buddolla V. (Salt Lake City, UT: Academic Press), 307–319. doi: 10.1016/B978-0-12-816328-3.00022-2
Raittz R. T., De Pierri C. R., Maluk M., Batista M. B., Carmona M., Junghare M., et al. (2021). Comparative genomics provides insights into the taxonomy of Azoarcus and reveals separate origins of Nif genes in the proposed Azoarcus and Aromatoleum genera. Genes 12 (1), 71. doi: 10.3390/genes12010071
Rognes T., Flouri T., Nichols B., Quince C., Mahe F. (2016). VSEARCH: a versatile open source tool for metagenomics. Peer J. 4, e2584. doi: 10.7717/peerj.2584
Seitzinger S. P. (1988). Denitrification in freshwater and coastal marine ecosystems: Ecological and geochemical significance. . Limnol. Oceanogr. 33 (4), 702–724. doi: 10.4319/lo.1988.33.4part2.0702
Shukla M., Arya S. (2018). Determination of chlorice ion (Cl-) concentration in ganga river water by more Mohr method at kanpur, India. Green Chem. Lett. Rev. 4 (1), 6–8. doi: 10.18510/gctl.2018.412
Siebel E. A., Kott A. E. (1937). Determination of specific gravity. J. Assoc. Off. Agric. Chemists. 20 (3), 535–542. doi: 10.1093/jaoac/20.3.535
Soltanpour P. N. (1985). Use of ammonium bicarbonate DTPA soil test to evaluate elemental availability and toxicity. Commun. Soil Sci. Plant Anal. 16 (3), 323–338. doi: 10.1080/00103628509367607
Su W., Zhang L., Li D., Zhan G., Qian J., Tao Y. (2012). Dissimilatory nitrate reduction by Pseudomonas alcaliphila with an electrode as the sole electron donor. Biotechnol. Bioeng. 109 (11), 2904–2910. doi: 10.1002/bit.24554
Trivedi P., Leach J. E., Tringe S. G., Sa T., Singh B. K. (2020). Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18 (11), 607–621. doi: 10.1038/s41579-020-0412-1
Vandieken V., Mußmann B., Niemann H., Jørgensen B. B. (2006). Desulfuromonas svalbardensis sp. nov. and Desulfuromusa ferrireducens sp. nov., psychrophilic, Fe(III)-reducing bacteria isolated from Arctic sediments, Svalbard. Int. J. Syst. Evol. Micr. 56 (5), 1133–1139. doi: 10.1099/ijs.0.63639-0
Von Glasow R., Jickells T. D., Baklanov A., Carmichael G. R., Church T. M., Gallardo L., et al. (2013). Megacities and large urban agglomerations in the coastal zone: Interactions between atmosphere, land, and marine ecosystems. Ambio 42 (1), 13–28. doi: 10.1007/s13280-012-0343-9
Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 73 (16), 5261–5267. doi: 10.1128/AEM.00062-07
Wang B., Lüttge U., Ratajczak R. (2004). Specific regulation of sod isoforms by NaCl and osmotic stress in leaves of the C3 halophyte Suaeda salsa l. J. Plant Physiol. 161 (3), 285–293. doi: 10.1078/0176-1617-01123
Wei Z. M. (2012). Preliminary research on dissimilatory nitrate reduction to ammonium in microaerobic condition (Guangzhou: South China University of Technology).
Yang C., Chen H. Y., Li J. S., Tian Y., Feng X. H., Liu X. J., et al. (2019). Soil improving effect of Suaeda salsa on heavy coastal saline-alkaline land. Chin. J. Eco-Agr. 27 (10), 1578–1586. doi: 10.13930/j.cnki.cjea.190178
Yan J., Wang L., Hu Y., Tsang Y. F., Zhang Y. N., Wu J. H., et al. (2018). Plant litter composition selects different soil microbial structures and in turn drives different litter decomposition pattern and soil carbon sequestration capability. Geoderma 319, 194–203. doi: 10.1016/j.geoderma.2018.01.009
Yuan Z., Druzhinina I. S., Labbé J., Redman R., Qin Y., Rodriguez R., et al. (2016). Specialized microbiome of a halophyte and its role in helping non-host plants to withstand salinity. Sci. Rep-UK. 6, e32467. doi: 10.1038/srep32467
Zhang Y. H., Ding W. X., Luo J. F., Donnison A. (2010). Changes in soil organic carbon dynamics in an Eastern Chinese coastal wetland following invasion by a C4 plant Spartina alterniflora. Soil. Biol. Biochem. 42 (10), 1712–1720. doi: 10.1016/j.soilbio.2010.06.006
Zhang X. W., Ji Z., Shao Y. T., Guo C. C., Zhou H., Liu L. F., et al. (2020). Seasonal variations of soil bacterial communities in Suaeda wetland of shuangtaizi river estuary, northeast China. J. Environ. Sci. 97, 45–53. doi: 10.1016/j.jes.2020.04.012
Zhang H. X., Zhao J. X., Chen G. J., Du Z. J. (2016). Kordiimonas sediminis sp. nov., isolated from a sea cucumber culture pond. Anton Leeuw. Int. J. G. 109 (5), 705–711. doi: 10.1007/s10482-016-0671-z
Keywords: Suaeda glauca, bacterial diversity, functional rhizobacteria, biotransformation, muddy coast
Citation: An X, Wang Z, Jiao K, Teng X, Zhou R, Xu M and Lian B (2023) Bacterial community characteristics in the rhizosphere of Suaeda glauca versus bulk soil in coastal silt soil modified by sea-sand and their implications. Front. Mar. Sci. 9:1001449. doi: 10.3389/fmars.2022.1001449
Received: 23 July 2022; Accepted: 16 December 2022;
Published: 09 January 2023.
Edited by:
Milko Alberto Jorquera, University of La Frontera, ChileReviewed by:
Vyacheslav Vladimirovich Shurigin, National University of Uzbekistan, UzbekistanMuhammad Arif, Southwest University, China
Copyright © 2023 An, Wang, Jiao, Teng, Zhou, Xu and Lian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Lian, bin2368@vip.163.com; Min Xu, xumin0895@vip.sina.com
†These authors have contributed equally to this work and share first authorship
 Xiaochi An1†
Xiaochi An1†  Bin Lian
Bin Lian